Dear all,
We are working on 130 kDa soluble protein; recently collected a dataset and solved the structure using cryosparc v2.12.4. After Ab-Initio classification we ran homogeneous refinement and observed a hugh dip with the corrected mask parameter (in the FSC curve; see the pic), although the resolution was estimated as 5.4A, but the map doesn’t look like that.
What possibly we have been doing wrong?
Thanks
E.R
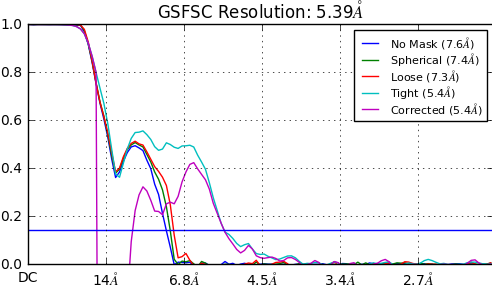
1 Like
Generally if the purple line (corrected FSC) and the light blue line (tight FSC) diverge, this indicates that the (dynamic) masking used during refinement was too tight and the FSC curves are therefore not trustable. At these resolutions it may be that your data quality is not great - do you see evidence of preferred orientations? How do 2D classes look?
1 Like
How can you make (dynamic) masking not so tight?
@yurotakagi you can set the dynamic mask “near” and “far” parameters in units of Å that control the tightness of the dynamic mask. You can also adjust the threshold at which the starting volume for the dynamic mask is generated.
1 Like
Hi,
I am facing the same dip in FSC while running non-uniform refinement.But map which is generated make sense.It is showing resolution of 4.5 angstrom.The dynamic mask is cutting density of map.Then,I tried changing dynamic mask far and near parameter.This didn’t help me.Can you please suggest what parameter I should try.
Hi,
Can someone suggest me for the same?It would be really helpful.
