Hi!
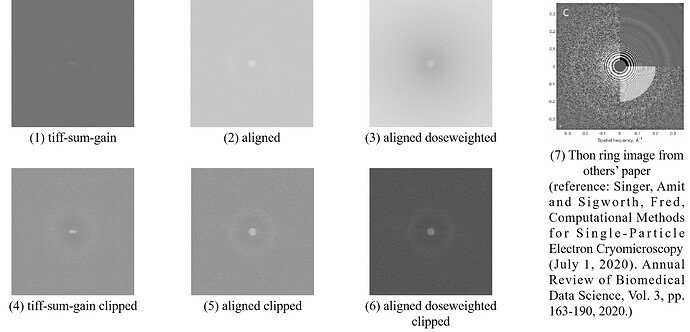
I’m trying to get Thon Ring images of micrographs using several lines of python code, but the results are not satisfying. As shown in following figures, compared to Thon ring image in published paper (figure 7), my results (figure 1 to 6) are worse and Thon rings are not so obvious. Is there any image processing step missed or wrong? Do you have any suggestion?
Here are details. The data used is from cryosparc tutorial T20S data (EMPIAR 10025 subset).
First, I read in the raw movie .tiff file, and sum over frames using numpy.sum(). Then, I read in the gain reference .mrc file, and apply it to summed micrograph by element-wise multiplication. After that, Fourier transformation is perform to the micrograph using numpy.fft.fftshift(numpy.fft.fft2()). Finally, I calculate the amplitude, use Logarithmic function to scale the values with 20*numpy.log(numpy.abs()), and display the result. This results in figure 1.
Meanwhile, I try to get Thon Ring images from motion corrected micrographs as well. In the results of cryosparc motion correction job, there are several different files and information for each micrograph (by the way, I’m not so sure what each item means, does any one know?), and I read in both aligned and aligned doseweighted .mrc file. Then, I use numpy.fft.fftshift(numpy.fft.fft2()) to perform Fourier transformation and 20*numpy.log(numpy.abs()) to calculate amplitude. The results are figure 2 and figure 3, respectively.
What’s more, I also notice the contrast_normalization function mentioned in another post (https://discuss.cryosparc.com/t/differences-between-2d-ctf-when-viewed-in-cryosparc-vs-exported-ctf-diag-image-path/10511/6). So I apply this function to figure 1 to 3, and results in figure 4 to 6, respectively. However, all my results have big difference with figure 7, whose Thon ring is more obvious and the entire figure looks more correct.
I don’t know why is the difference. Probably some image processing steps are missed or some steps are wrong? Any one has any advice? Thanks!
Best,
Ciren