Hi all, Tthe tutorial indicates that templates we select from the 2d classification should be at the center of the box so that we can get more accurate coordinates of particles at template picking.
But when I select the proper templates in select 2D job, I found the result of the 2d classification is not ideal. There is not template at the center of the box.
I tried different diameters of the blob pickers, different iteration numbers of 2d classification, but the results are still not ideal.
Does anyone know the possible cause or solution? Thanks in advance!!
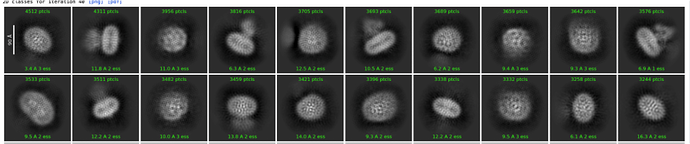
This is fine, class 2,4,6,10 are normal. The detergent belt is center of mass, and this should be fine for template picking. If you really want POI in the middle, there are several ways to do it. Load mask, run local refine, the early info of that running job tells you the mask location, then can use those coordinates to recenter the particles. Run 2D again with no recentering
1 Like
Thank you so much! Your suggestion does help.
Hi! I have two more questions that I would appreciate if you knew the answers! How to identify the top view classes as class 2,4,6,10 are all the side views of the protein? Or, does the top view matter to template picking? Besides, does the resolution of the class averages (template) matter to template picking? Namely, is it neccessary to do double 2d classification to get clearer class averages? Thank you in advance!!!
Do not need high-res clean templates for template picking. But if you run Topaz, then yes you want to run 2D again and select only the best particles.
I would assume most other templates are top/bottom views and yes, you do want to pick them as well. If you get a good 3D model or ab initio, then you can run “create templates” job and it will show you what all the views would look like.
Others are more expert at this, I have never worked with membrane sample
Thanks so much for the information!
Yes, resolution does matter for template picking, but not in the way you think - to avoid as much bias as possible and “Einstein from noise”, you actually want to deliberately limit resolution, which is why templates and micrographs are low pass filtered (by default to 20A). See this thread for a couple of posts with some explanation of why high resolution picking is so dangerous.
Not sure whether your sample is supposed to be a monomer or dimer but it looks like both are present (or there are some “twins” in the same detergent belt, different scientific relevance, same overall effect) but I’d probably take classes 10 and 11 (top right and bottom left respectively) and template pick using just those. You’ll find that due to the low pass filter, it’s not very “specific” and will still likely pick all of the smaller densities, but if not try picking with classes 1, 2 and/or 12 as well.