Hello @hai!
I think it may be worth considering whether the particle you are looking for is truly present in the micrographs. @rbs_sci’s point here Blob pick does not pick the right particles - #4 by rbs_sci is an important one — if you provide high-frequency templates during template picking, you run the risk of template bias.
There’s a rather famous example of this phenomenon here: https://www.pnas.org/doi/full/10.1073/pnas.1314449110, which reproduces the also-famous “Einstein from noise” figure from an earlier paper on model bias. In the PNAS paper, Richard Henderson discusses how carefully picking “particles” which are in fact pure noise can create convincing-looking 2D class averages, even if the particle is not present at all.
I will also provide a concrete example here:
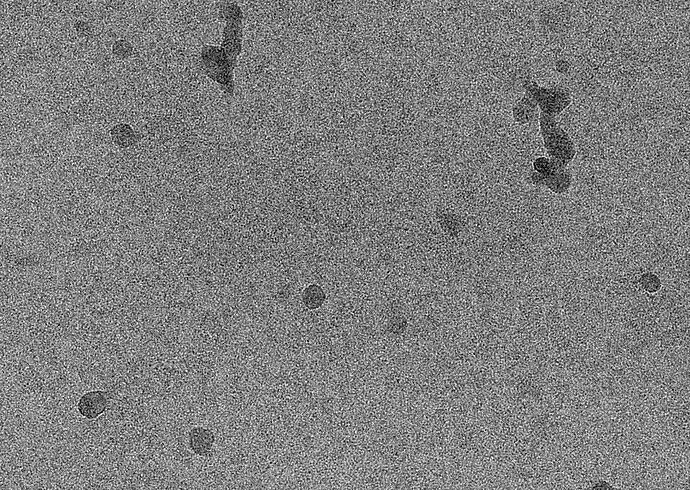
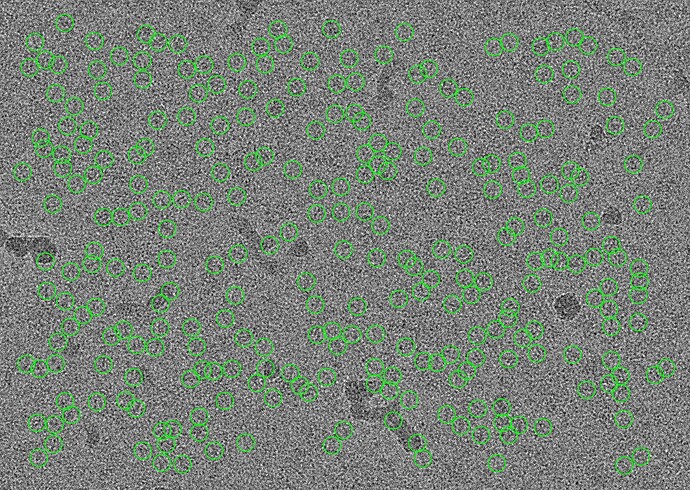
Let’s take a look at these beautiful micrographs from EMPIAR-10424. These are micrographs with lots of high-contrast ApoF particles in them:
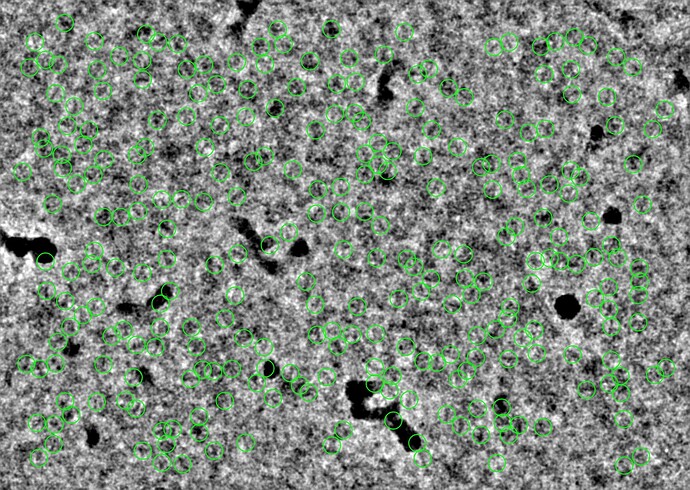
But let’s say I’m looking for a different particle, one that looks like the Structura logo. I’ll provide this template to Template Picker, and set both lowpass filters to 1 Å.

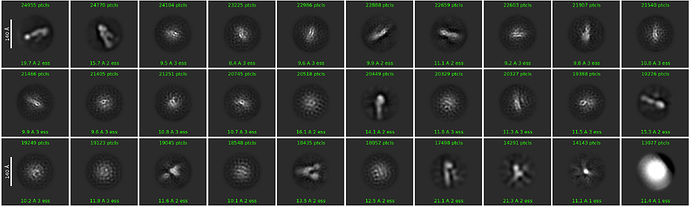
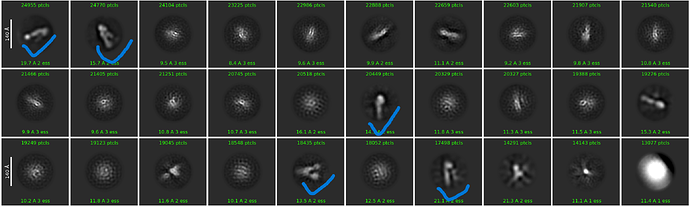
In essence, when you perform template picking, you are asking CryoSPARC to look through every position of your micrograph and find patches which look like the templates. In my case, even though there are clear ApoF particles and certainly no visible particles which look like the Structura logo, I still found 39k particles! And when I average those particles together, I get a “2D class average” which looks convincingly like the logo:
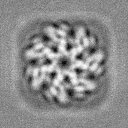
Of course, we know that there are no particles in these images which look like the logo. Instead, Template Picker picked thousands of positions of noise which, by random chance, happened to look a bit like the template we gave it. Each one individually doesn’t look like much at all, but averaged together the trend emerges.
It’s very important to avoid this effect, so the field generally recommends filtering your templates to a very low resolution. The default in CryoSPARC is 20 Å, but when you are providing data from outside your project you cannot be sure that such a template is present in your data at all. In these cases, it is necessary to filter the templates even more aggressively. This is why @rbs_sci suggested increasing the filter value to 30 Å in your case.
If you could find your particles using the blob picker, then you could be more confident that they are actually present in your micrographs. However, it would still be important to lowpass filter the templates, since you can pick biased noise even if the particles are present! In general, it is rarely correct to reduce the filter value of the template picker.
I’m sure this is not the answer you were hoping for, but I think in this case I would be very wary of trusting the template picker results until you can produce class averages which look like your particle using a less-biased technique, such as blob picking.
I hope that is helpful!