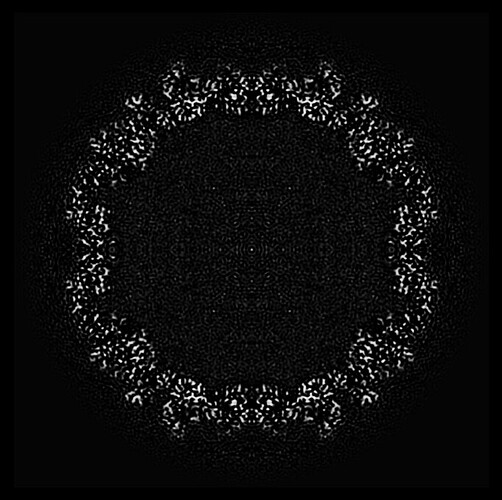
This is a cross-sectional view of the 3D structure of the virus I constructed. From the exterior, it appears to be a regular icosahedron with a diameter of around 300 Å. It belongs to symmetry I, and its interior consists of RNA and proteins.
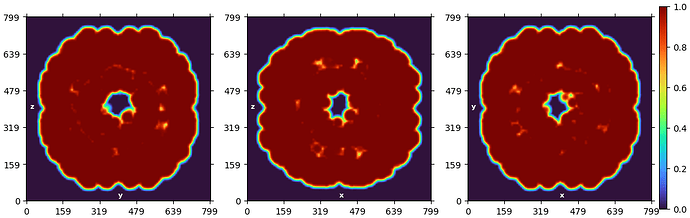
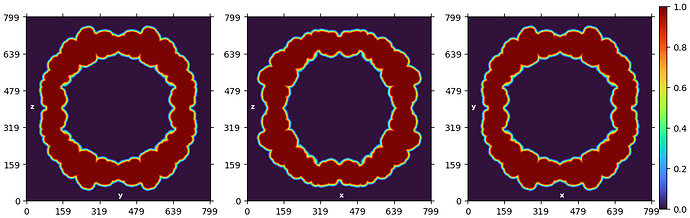
This is the mask I used when constructing the 3D structure, which was generated by 3D classification.
Due to the absence of RNA content in some viruses, a mask excluding the central structure was generated during 3D classification.
However, I believe the internal content is not symmetrical, so I want to mask out the outer layer and analyze the central content separately, which has a diameter of approximately 250 Å. Therefore, I would like to ask for guidance on how to mask out the surrounding layer.
Hello @Barb!
I will start by saying that I do not have much experience with viruses — others in the forum will likely have more to add to what I say here. However, I can hopefully still point you in the right direction!
Particle subtraction
In your case, you have a large, well-aligned exterior and an interior of unknown quality and symmetry. In this case, I’d reach for Particle Subtraction.
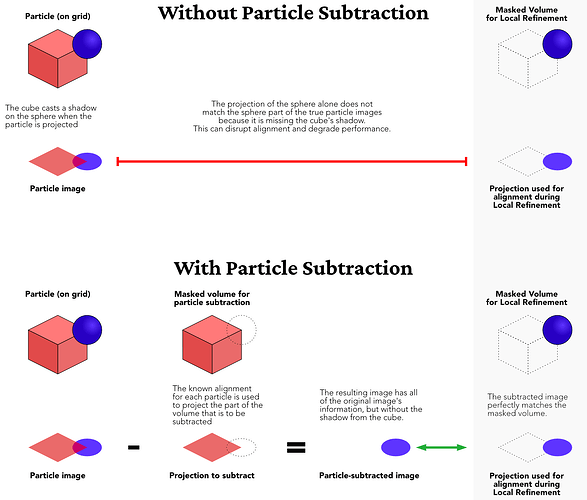
In this technique, we first make a mask around the part of the target we want to ignore. It looks like you’ve already done that, with your second mask. We use this mask to create a projection of just the part we want to ignore in each image’s pose. We then subtract that projection from the image. This will hopefully leave us with images just of the other parts of the target, that is, the parts that were not inside the mask used during Particle Subtraction.
Here is an explanatory image from the above guide page to help illustrate this process:
There is also a section of our Local Refinement guide that goes into more detail on how to set up a Particle Subtraction job and use it in combination with Local Refinement to improve the quality of small or otherwise poorly-aligned regions of a map.
Symmetry Relaxation
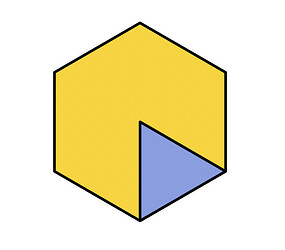
Another technique you might try is Symmetry Relaxation. This is more likely to work if the internal part of the virus is itself rigid, but is poorly resolved because it has a different symmetry order than the external part. For a 2D example, consider this shape:
The outer part is C6 symmetric, while the inner part is only C1 symmetric. However, the inner part will always be the same triangle. In a case like this, Symmetry Relaxation improves the inner map by checking all six symmetry-related positions and using the one that puts the inner triangle in the same position for each particle. There are more real-world examples in the guide page.
I hope that’s helpful!
Hi, rposert!
Thanks so much for your suggestion
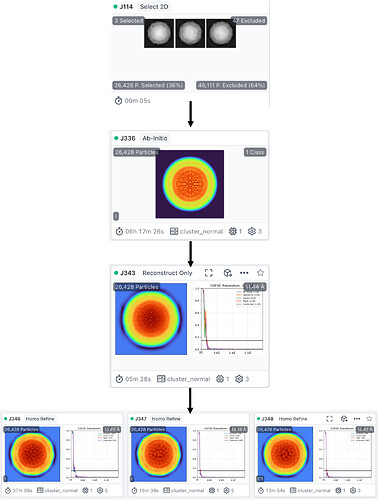
I tried Symmetry Relaxation, but I’m not sure what went wrong, and the final resolution was very low.
Hi @Barb!
A slight decrease in GSFSC resolution is expected when you turn on symmetry relaxation. When we enforce symmetry, each particle is used N times (in the case if icosahedral symmetry, 60 times!), once for each symmetry position. However, symmetry relaxation only uses the particle once in the pose it fits best. Therefore, your overall signal-to-noise ratio decreases when turning on symmetry relaxation. However, if there is asymmetry present, the overall quality of the map will generally improve, since the particles are not truly symmetric.
As for this workflow — is there a reason you never perform Homogeneous Refinement on the virus? Does your map from J343 have the expected symmetry?