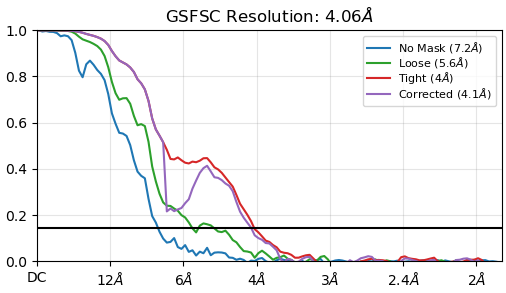
Hello everyone, I am currently analyzing the structure of a protein with a molecular weight of about 90KDa. After Ab-Initio, Homo-Refine and Nu-Refine, the values obtained so far are as follows. Do you have any suggestions for further improving the resolution? Since the number of particles calculated is only 28,304, how can I increase the number of good particles?Thanks.
From how many micrographs?
It’s kind of stating the obvious, but the easiest way to increase the number of particles you’re working with in collect more data.
For a 90 kDa complex, at 4 Ang, can you see believable features in the helices? Beta sheet strands clearing up?
Otherwise, try Local and Global CTF refinement?
Hi, we collected more than 5,800 pieces of data, and we were able to see some of the protein signature structures.
We collected more than 5,800 pieces of data, and I wondered if we could find more usable particles in the existing data? Thank you.
Repick using templates created from your 4 Ang 3D refinement?
To be more specific, more information is really needed. And sometimes you just have to go back and collect another dataset. Other times, you have to do the best you can with what you have.
OK, thank you for your advice.
If you are re-collecting try on HexAuFoil (yours is small enough) or UltrAuFoil.
Does it really look like a 4A map ?