Hey all. I can’t get rid of a certain graininess in my averages. I have tried to change the reconstitution resolution but this didn’t change much.
Is it due to bad image quality in the first place or are there other parameters I could change in the 2D classification job?
Hi Dario, likely signal to noise.
see Streaky classes for some answers.
Thanks a lot, I will go and try the suggested answers.
Hi @DarioSB! Thanks for posting here!
@ehanssen is right, the streaks you saw in your first 2D classes are typically due to overfitting of noise instead of your particle. The solutions presented in the linked discussion are both ways of preventing this overfitting. I’ll explain a bit why those settings help in situations like this!
Turning off Force Max allows cryoSPARC to essentially “smear” the particle across a few different orientations and shifts when it’s not certain about the alignment. This helps prevent noise from piling up when alignments are uncertain, but also makes the process take longer since those smears have to get narrowed down.
To explain the second part of the solution (increasing the final full iterations) we need a bit of background on how 2D classification works. Rather than use all of the particles for every iteration, cryoSPARC uses a smaller subset (a “batch”) of particles for each iteration. This really speeds up the processes of generating your 2D classes!
For large particles this works fine, because there are lots of features to align. For smaller particles, though, this can create a situation where there is not enough information in a single batch to improve the classes, so things start to get noisy. To fix this, you can do a few things.
- First, you can increase the number of particles used in each batch (
Batchsize per class) so there’s more information to use during each iteration. - You can also increase the number of times cryoSPARC goes through the full particle set (still in batches) by increasing
Number of online-EM iterations. - Finally, you can increase the number of times cryoSPARC uses the whole dataset at once (i.e., no batches). This is what
Number of final full iterationsdoes. These no-batch iterations are very slow, and that’s why the default is set to 1.
I hope that helps explain why those settings improved your 2D classifications!
Now, as for the results you posted with those settings changed. Can I ask what about them makes you suspect they are overfitted?
I also wonder if I can see what all of your 2D classes look like — I’m curious if you have a large number of particles in junk classes, for instance.
Hmm, I agree, these do look a little strange. May I ask you a few more questions?
- How many micrographs are these particles from?
- How many total particle picks did you start with, before doing any 2D classification or cleaup?
- About how big do you expect your particle to be (in Å)?
- How big is your extraction box size for these particles (before downsampling; in Å)?
Thank you! Hopefully we’ll be able to figure out what’s up here!
@DarioSB May i know the size of the protein? I assume that these particles are small for which you are facing this issue. I am having the same problem and currently its unsolved. I would recommend you to use neural network based algorithms for picking and extensive cleaning at the step of ab initio and heterogeneous refinement. Everything that @rwaldo mentioned should give you good classes, but you should also have sufficient directional views to have a successful reconstruction.
@rposert
Thanks, here are some more details:
- I started with 4500 micrographs, after curation there were 737 left that I considered to be usable.
- From these 737 mics I extracted 280k particles from a template picking job.
- After 3x “2D classification” / “select classes” I ended up with 46k particles shown in the image I posted
- The particle is 120kDa and should be around 120Å at the longest side (at least the part that is shown, there is another flexible part that I wont be able to resolved)
- The extraction box size is 250px (=275Å) and I didnt use any downsampling
@Das
about 120kDa. I start to doubt that it is the picking itself since the picking jobs seem ok, they seem to pick actual particles.
Thanks for that info @DarioSB!
-
Am I correct in understanding that you had to discard almost 85% of your movies? That’s too bad! Could we see a few examples of the micrographs you kept and those you discarded?
-
That is a pretty good particles/micrograph! Probably at the higher end of the distribution. I have a few questions about this as well, if you don’t mind:
a. How did you generate the template for template picking?
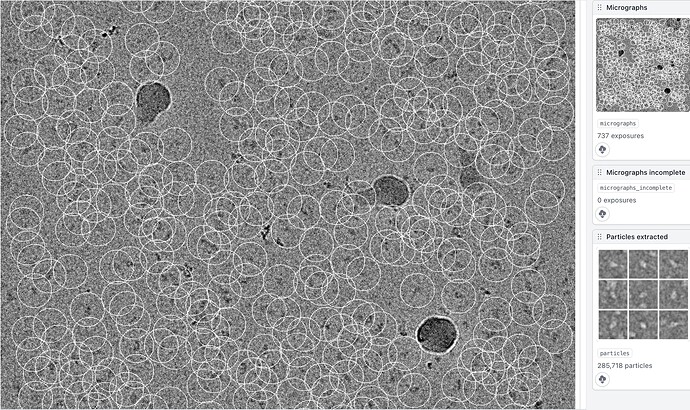
b. Could I see the thumbnail with the particle picks? The easiest place to find this is probably actually the Extract From Micrographs job, if you click the
Show From Topbutton in the top-left and then scroll down a bit and find the image with your micrograph and a bunch of white circles.
Inded @rwaldo, I had to discard a lot of data due to thick ice unfortunately.
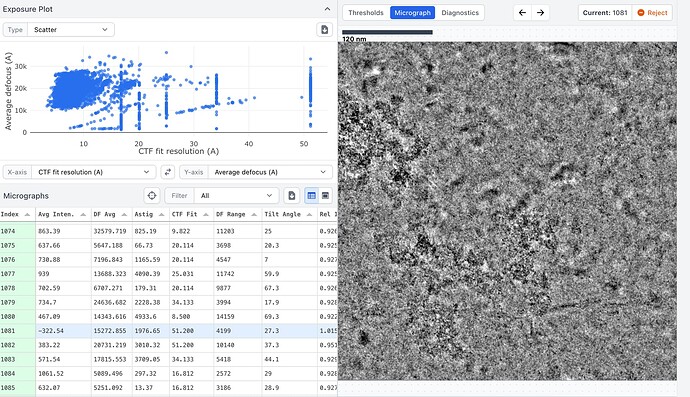
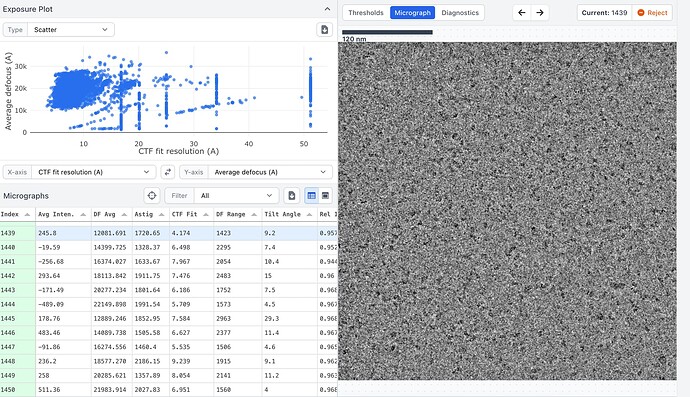
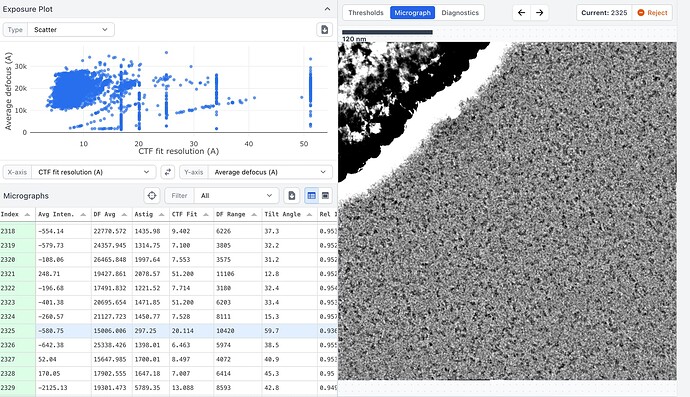
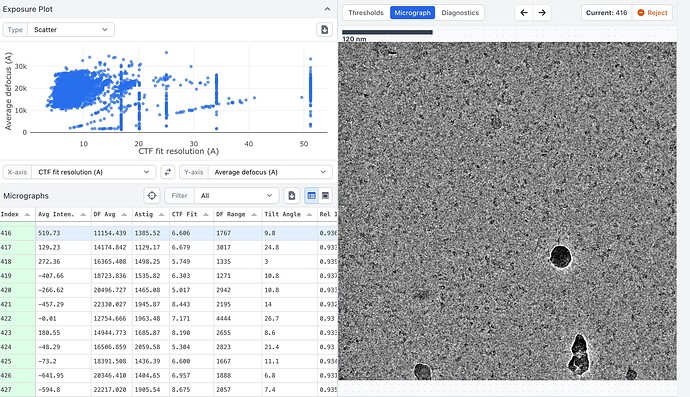
Here are some example mics, one with bad CTF fit and one with better values. Also one with low and one with high defocus
Here is the picking thumbnail. The template was created by a first round of 2D classification, so nothing external but only from the data itself
These classes don’t look too bad to me given the number of particles per class and the size of the particle. You may want more O-EM iterations though - Class2D is very slow to converge when Force/Max is off.
I already have 30 O-EM iterations. How many do you suggest?
40 usually, sometimes up to 80