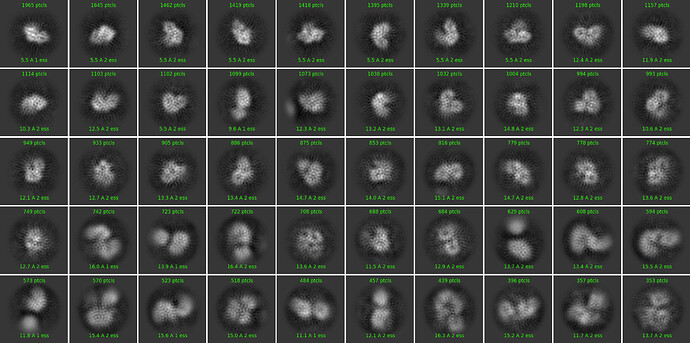
hi,
i have a small protein ~70kDa with a good movie data set. however the 2D classes have radial streaks. I have tried everything I could think of to avoid over fitting: decrease the alignment resolution, enforCe non negativity/clamp solvent, Increase the batch size …
Looking for new ideas, or feedback on the classes.
Maybe the region you collected are too thick for your sample. You may need to collect another thin ice dataset.
Hi Eric,
A couple of things to try (either alone or together):
- Switch Force max off to marginalize over poses and shifts (and increase the number of online-EM iterations to 40). Often helps with small proteins, but takes much longer to converge (hence the increased iterations). Increasing the batch size to 400 often helps in combination.
- Increase the box size - this often helps with very small proteins.
- Try using a much larger number of final full iterations - say 20 - with force max on. This will take a long time, but can sometimes give much improved classes.
Hope that helps!
Cheers
Oli
we are as thin as the particle want to go in (most micrographs have part of thm with no particle to make sure we are as thin as it is possibel to be)
Oli, it seems to work to some extend, not as good classes as i would like with the quality of the dataset but much improved. thanks.
That’s the problem I have met. Some kinds of proteins don’t like staying in thin ice. We still can’t solve this problem for some small proteins.
Based on my experience, we have determined a 80 kDa membrane protein structure, but it looks very similar like your case when we tried to do the same thing for the same protein of another species. We used the same condition to prepare these two samples, the only differences are species and collecting region.
So I believe this is the problem about the ice, even though we collect micrographs as thin as we got.
i probably would agree that the thickness of the buffer is too much.
one thing from Oli, about the box size, not sure if it is teh right forum but nonetheless i will ask. Why are smaller particle better off with a bigger box size?
Yes - larger box size relative to the particle size - because displacement of CTF information from the particle includes an absolute offset which is dependent on defocus and electron wavelength but not on particle size, see this CCPEM thread and reference therein for discussion:
https://www.jiscmail.ac.uk/cgi-bin/webadmin?A2=CCPEM;47cdfca4.1803
Result is that if your particle is really small, the rule of thumb of twice the particle diameter won’t be adequate. The other implication is that data collected with more defocus will require a larger box size.
Cheers
Oli
yes i see what you mean. i usually calculate roughtly the size needed for the defocus applied and the particle size and add a bit more. what worked on the trick you gave me is the Switch force max off with a batch of 200 (400 gets worse somehow).
i misread teh comment on SFMO and thought on was for small particles. my english is lacking …
both are for small particles! and yes the batch size can require some tuning