Hi all,
I am processing a small dataset of a ~185 kDa protein complex with long flexible linkers using cryoSPARC. The visible domains may only account for ~120 kDa.
Several issues have arisen:
First, the particle distribution is very crowded. Even after reducing the protein concentration to 2.5× (0.15 mg/ml), the sample remains overcrowded.
I manually picked particles for Topaz training, and the resulting Topaz picks appeared well-centered. However, some white dots around the particles were still visible, likely due to neighboring particles.
During 2D classification, the resulting classes consistently had a grey background and appeared very blurred.
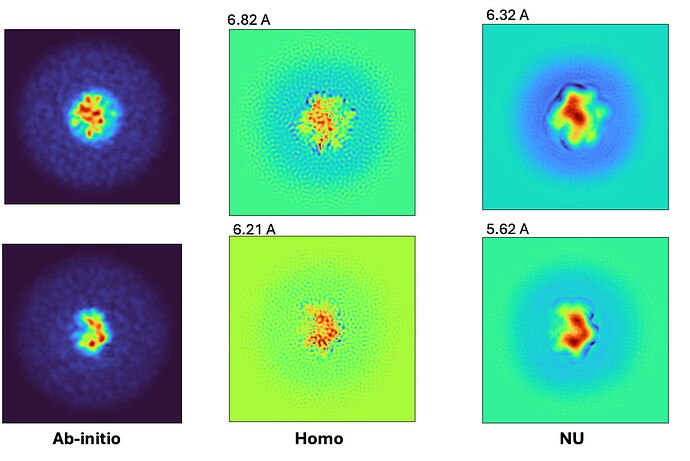
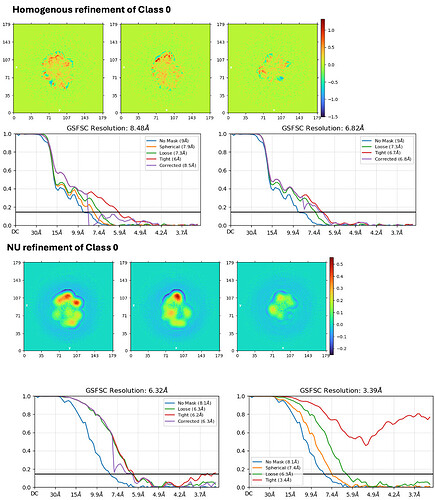
In 3D refinement including heterogenous refinement, homogenous refinement, and NU refinement, they all got worse resolutions compared to the reconstructions generated from ab-initio, especially in NU refinement.
Here is some details of my data processing workflow:
Import: 1.65 A(pixel size) , 200 kV, 33.06 e/A^2, 30 frames, 3710*3838
Patch motion corr. → Patch CTF (172 micrographs)
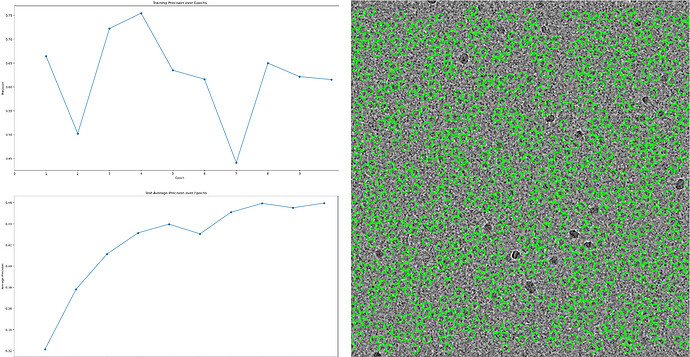
*Manual picked 434 particles for 2D classification, 390 particles were selected for topaz training.
After iterative rounds of topaz training, 139,239 particles were picked and extracted with a box size of 168 pix, bin to 84 pix.
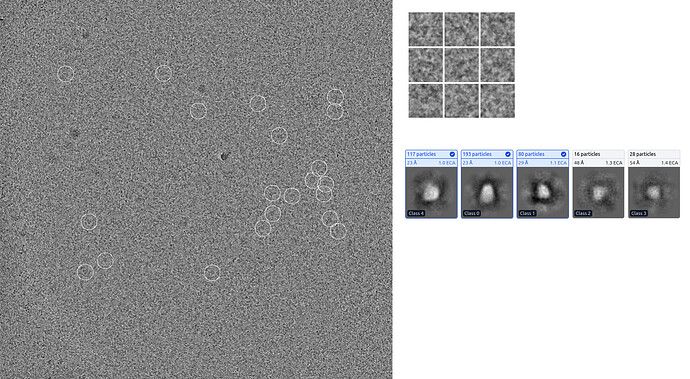
2D classes, Mask diameter: 140Å :
They were all fuzzy with grey background even after I increased the batch size to 300 and iterations to 50.
The best 2D classes I could get after several rounds of picking and 2D classification :
With the use of “enforce the negativity” in 2D classification, I got these classes with the total particle number of 202,255:
I threw these particles into ab-initio with 2 classes.
Then I did 3D homogenous refinement followed by NU refinement of each class.
The map resolutions from 3D refinement jobs all look worse than that from ab-initio even though the numbers are better.
I’m still relatively new to cryo-EM data processing, and I’m wondering whether there might be something wrong with my workflow, or if there’s anything I can do to improve the current results.
Specifically, I’m curious: what kinds of issues can lead to grey backgrounds and fuzzy 2D classes during classification?
And also, what kind of problem can lead to worse resolution in 3D refinements compared to ab-initio?
Please let me know if I need to provide further information.
Thanks.
Best,
Jessi