Hey everyone,
I am new the cryoEM/SPARC and would appreciate any help I can get. I am working on a protein that is ~125kDa as a monomer, and 250kDa as a dimer. The dimer is an elongated structure (have crystal structure), while the monomer is globular (no structural information). I am looking at discerning both structures which should be present in the data collected, although the dimer will likely contribute to the majority of the particles it can be induced not only from ligands but also concentration.
I have collected a dataset of 1.3million particles and have done the following general processing.
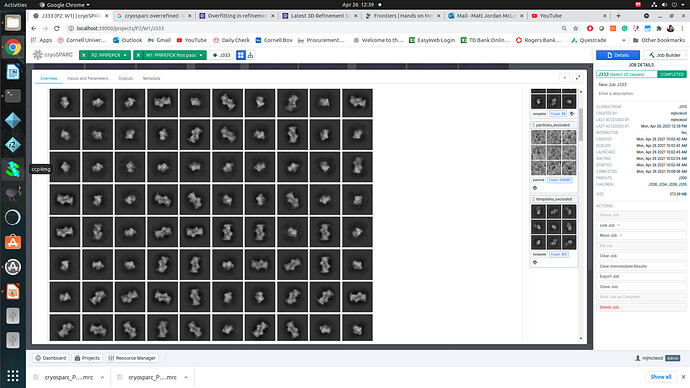
Started with blob picker, 2D classification, (followed by generous selection) and ab-initio to get ~ 3 classes (dimer, monomer, and some “Junk” reconstruction). I used these inputs to create templates and particle pick. From the new 2D classification (300 templates - 3 rounds to clean up classes) and curation I have approximately 800K particles left over. I have both selected generously and stringently (with high resolution and low ECA cutoffs ~350K particles) followed by ab initio for two classes. These two reconstructions look as expected, there is a high agreement with the dimer crystal structure and the volume of the monomer is consistent what what I would expect.
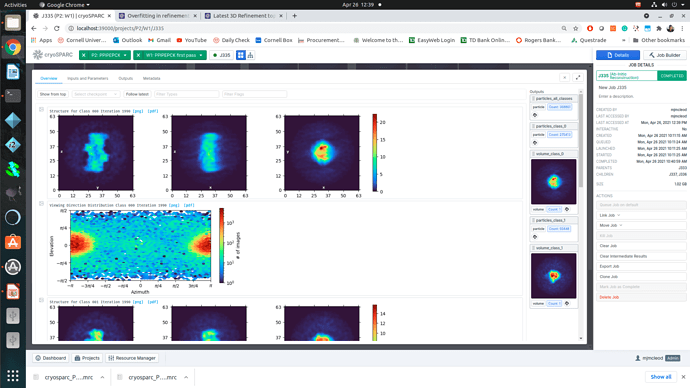
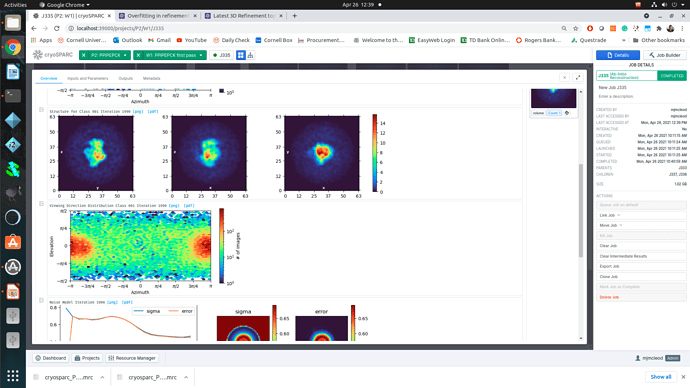
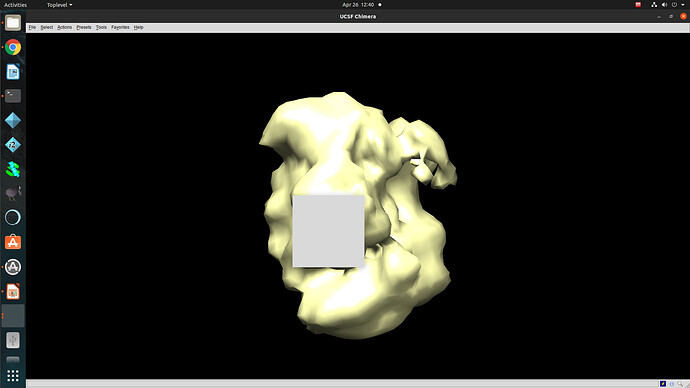
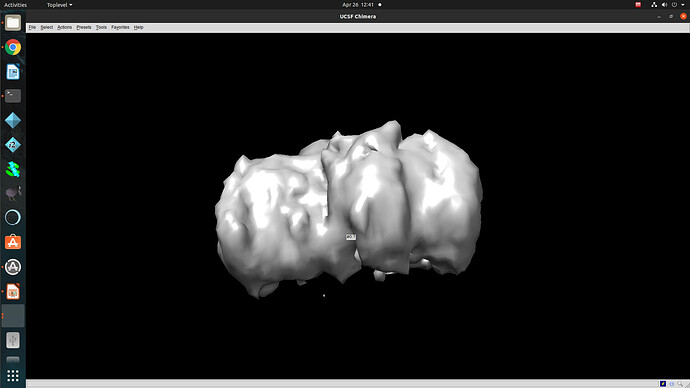
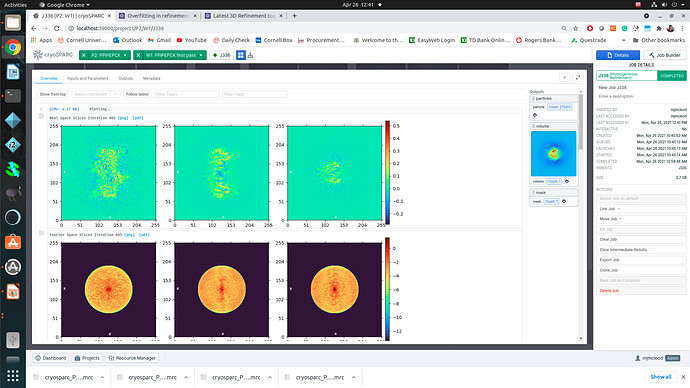
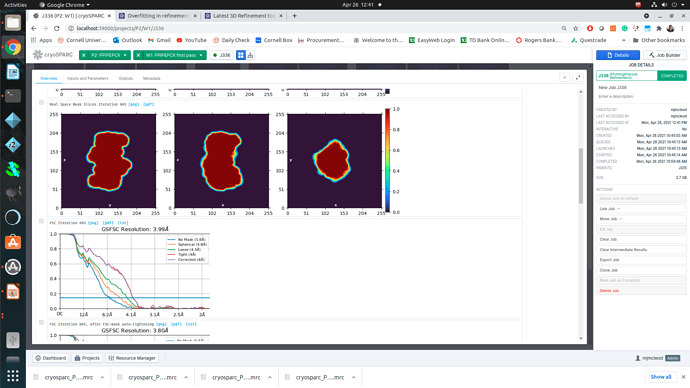
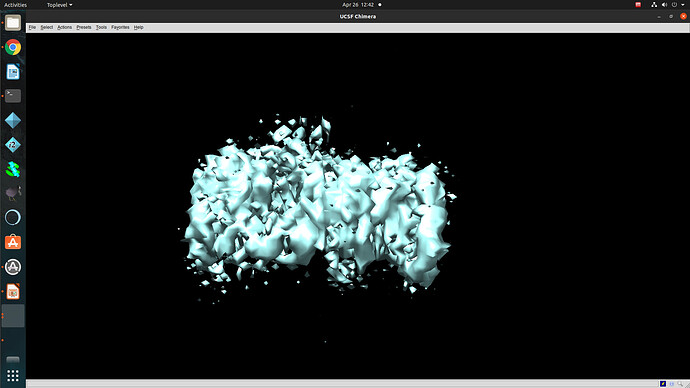
Regardless of how I treat the data upstream of refinement, the refinements essentially all turn out the same. This would suggest to me that I am doing something wrong or there is a pathological problem with the data (orientations). The refinements have high CC until 12A, where there is a significant drop to ~0.6CC followed by a slow decay to approximately 4A. I assume that there may be some high resolution data present but the medium resolution data is missing or is washed out (poor alignments or?). After refinement the maps look awful, with no high resolution structural data and protrusions around the volume.
I am unsure if there is an orientation problem (probably likely), or if the refinement is having trouble with this dataset and it is potentially fixable. From what I have gathered from the forum, small particles reconstructions can suffer from over-refinement which may also be occurring here. Any insight would be appreciated! I am happy to supply anything that can help.
 thanks alot!
thanks alot!