Hi, I am new to cryoEM and cryoSPARC and attempting to process my first dataset. I am running into some problems I am hoping someone can help with. I’m working with a relatively large dataset of a non-symmetrical complex with approximately 200-120 A dimensions. The complex oligomerizes, forming dimers and trimers, with monomers and trimers being the most common species. Additionally, the dataset is a bit dense with frequent particle overlap which must be weeded out from pure monomers or actual oligomers. The trimer appears to exhibit preferential orientation within the ice, so I am now focusing on resolving the monomer instead. Initially, I ran into two main issues with the monomer: 1) overlapping particles getting sorted into good looking monomer classes but with a soft cloud appearing in the area of the overlap, and 2) 2D junk classes getting averaged into good classes by the final iterations. I seem to have resolved these problems by turning off re-centering of 2D classes, masking, and the annealing of sigma. However, following ab initio reconstruction, I am having issues with hetero refinement FSC curves getting cut off. Per the forum, this can occur with downsampling, but I have not down sampled (because I have yet to figure out how to successfully do this ![]() . Here is my process so far:
. Here is my process so far:
Un-tilted data was collected on Krios with the following parameters:
accelerating voltage (kV): 300
spherical aberration (mm): 2.7
total exposure dose: 50.87
pixel size (A): 0.656
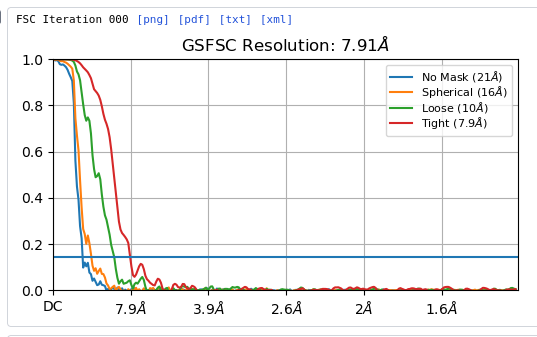
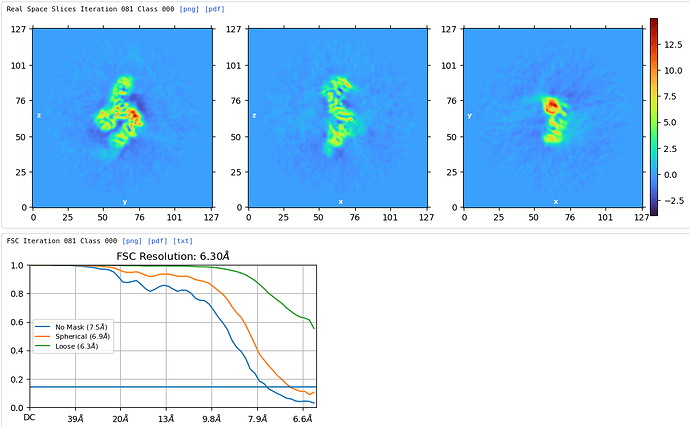
Patch CTF job was performed on a total of 15202 micrographs using standard parameters. An initial quick pipeline was used to generate 2D templates from a volume map, and a subsequent template picker job picked 6.1 million particles using standard parameters except for particle diameter (A) set to 220. A curate exposures job was used to pare micrographs down to 12,514, and a subsequent extract micrographs job using an extraction box size of 600 pix extracted 3.8 million particles. Seven rounds of 2D classification with standard parameters except: use circular mask on 2D classes = OFF, re-center 2D classes = OFF, Iteration to start annealing sigma = 200 (essentially OFF), number of final full iterations = 5, number of classes = varying from 200 to 80, number of online-EM iterations = varying from 40 to 30, generated 463,000 good particles. single class, 3 class, and 6 class Ab Initio was performed on just 100,000 particles to speed up processing time, and a heterogeneous refinement job was performed on the best classes but with the total 463,000 particle dataset. The volumes from these hetero refinements look great, but the FSC curves approach the nyquist limit and are cut off. I started a homogeneous refinement job on the single class ab initio and so far the FSC curves look normal.
I am wondering what I can do to improve my process and troubleshoot the wonky FSC curve. Thanks!
2D classes used for Ab Initio:
FSC cut off during hetero refinement
FSC look normal at iteration 000 from homogeneous refinement: