Hi all,
I am dealing with membrane protein particles without downsampling. The FSC curves from NU refinement look like quite strange that all the curves stick together. BTW, the particles were cleaned and the 2D result is very beautiful.
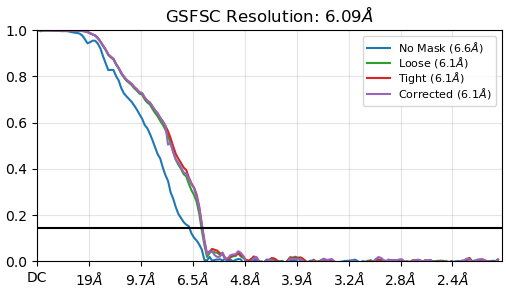
The ideal resolution of this dataset should go to 4A. Also any comments on that ?
When you say the ideal resolution should go to 4Å, what is that based on?
Do you see clear secondary structural features in the map, and does the fold match what you expect from 2D and prior knowledge about the protein?
Hi, @olibclarke. I have another dataset of this protein with similar particles which generated a final reconstruction at 4Å resolution. And the results of 2D classification of those two dataset are quite the same. Just to clarify, whether this issue can be resolved through data processing alone or if new data collection is necessary?
If it is a sample you have collected on before, 2Ds look ok and picking looks ok, I would suspect either an error during movie import (e.g. accidentally specifying wrong dose) or some severe optical aberration (e.g. excessive beam tilt). Have you tried running Global CTF? If so, what do the diagnostic plots in the log look like?
Thanks the suggestion from @olibclarke. No improvement after apply Global CTF refinement. The plots from global CTF are displayed below, are the the “diagnostic plots” you have mentioned?
Yes these are the plots, no evidence here of anything that would explain the low resolution. I would double check your parameters at import just to make sure that nothing went wrong there.
Well, the movies were motion corrected by MotioncCor2 outside of cryoSPARC and parameters at importing micrograph are just the “voltage:300, pixel size:1.087, spherical aberration: 2.7, Total dose: 50”. I would try to re-do motion or give it up if no improvement.
Sounds fine… the only other thing I can think of in the past which we have seen cause this is a shutter sync problem with the detector (where effectively you end up pre-exposing the sample). I would double check with the facility if users before or after you encountered similar issues, to rule out a hardware problem.