Features of proteins are missing after NU refinement. So, can we try masking and local refinement after homogenous refined volume which has more features than NU volume.
Hi @Shahsal! It’s too bad that you’ve lost some features from Non-Uniform Refinement. You can definitely perform a Local Refinement straight from a Homogeneous refinement, so your plan is sound.
Do you think you’d be able to share a few images of the features you noticed disappearing when you use Non-Uniform Refinement? I’d love to see if we can figure out a way for you to be able take advantage of it without losing information from your map!
Just to give brief idea about the protein.
We made domain swapped dimer of Fab fragment bound with antigen on two sides. But we are not able to define the complete map of antigen and see only 50-60% of features in 3D reconstruction (6 ab-initio models). Upon heterogenous refinement followed with homogenous which looses 10-15 % antigenic volume. But after NU refinement, it looses more features. Previously, we have solved this kind of complex with another dimer Fab and with same antigen and were able to build complete density of Fab-antigen along with glycans.
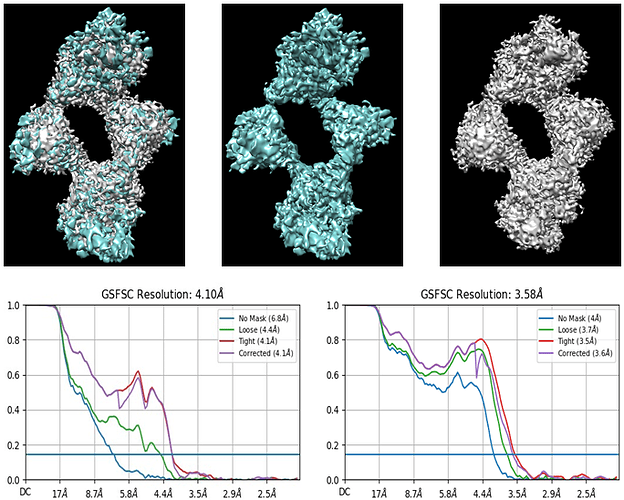
The image has overlay and individual volume of homogenous and NU refinement with their GSFSC curves.
Thanks for the images @Shahsal! I’m not used to looking at particles like this — do you think you could indicate where the antigen is, and which parts are the Fabs for me?
I see a few things here that I’m curious about:
The maps at this contour look pretty noisy to me. What do they look like if you contour them down?
I also notice on the right hand GSFSC plot (the one which goes to 3.58 Å) that the Corrected curve (in purple) never rejoins the Tight curve (in red). This typically indicates that the mask used for refinement is too tight, so it is introducing information to both half-sets, which can cause artifacts and noise in the final reconstruction. Have you tried any refinements with looser masks?