Hi all,
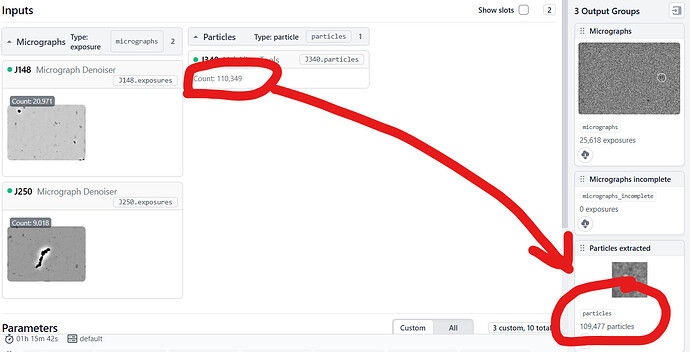
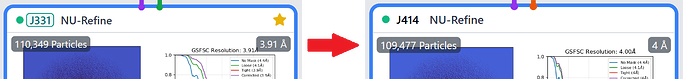
So, I have a strange issue. I extracted my particles from denoised micrographs using 512 px box size fourier cropped to 256 px. I ran 2D, ab initios, hetero refinement and subsequently NU refinement with approx 115k particles. Then I re-extract the same particles (connected particles from NU-refinement job and de-noised micrographs) but the extracted particles were approx 1000 short and this for some reason worsens the resolution of my map. How do I solve the issue? Secondly, I though using full size particles were supposed to yield better quality map than fourier cropped ones or am I wrong? Do I even need to extract full size particles? Please advise.
Re-extracting particles will lose you particles if you have Recenter using aligned shifts enabled and a box clips off the edge of the micrograph, as CryoSPARC does not “barcode” the off-micrograph box sides the way RELION does.
As for losing/gaining resolution, the real difference between 3.9 Ang and 4 Ang is rather moot and I doubt most would be able to tell a difference when examining the map alone. Further, unbinning particles is only 100% guaranteed to gain you resolution when you’ve hit the binned Nyquist limit, which from what little I can see of your 3.9 Ang FSC curve, you are nowhere close to doing. For your case here, I would not bother unbinning.
2 Likes
Thanks a zillion for clearing this out for me.
1 Like
No worries; good luck with the rest of your processing!