I wrote a script to pull in image shift data from .mdoc files generated by SerialEM, and cluster the image shifts into groups. Then I updated my exposures with that new group information, and saved it into my project. However, when I run the Exposure Group Utilities job, with those exposures, and check with “info_only”, it simply outputs a single group.
I assigned the groups to mscope_params/exp_group_id and verified by printing every row that the groups were being assigned (in my case, 0-24). I also loaded the output of one of my updated datasets, and checked that parameter, and it was indeed stored correctly.
Below is the CS portion of my script:
cs = CryoSPARC(license=cs_license, email=cs_email, password=cs_password, host=cs_hostname, base_port=cs_port)
assert cs.test_connection()
project = cs.find_project(cs_project)
job = cs.find_job(cs_project, cs_job)
cs_exposures = job.load_output(cs_job_output)
grouped_exposures = cs_exposures.copy()
for row in grouped_exposures.rows():
for i in range(len(exposures)):
if exposures[i]['exposure'] == row['movie_blob/path'].rsplit(sep='/')[-1]:
row['mscope_params/exp_group_id'] = exposures[i]['group']
break
#for row in grouped_exposures.rows():
# print(row['movie_blob/path'], row['mscope_params/exp_group_id'])
project.save_external_result(
workspace_uid=cs_workspace,
dataset=grouped_exposures,
type="exposure",
name="grouped_exposures",
slots=["mscope_params"],
passthrough=(cs_job,cs_job_output),
title="Grouped Exposures"
)
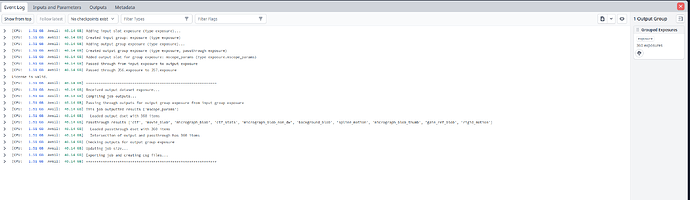
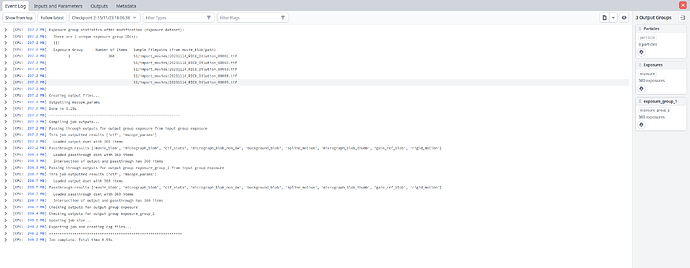
but they don’t seem to want to show up. Here is my external job:
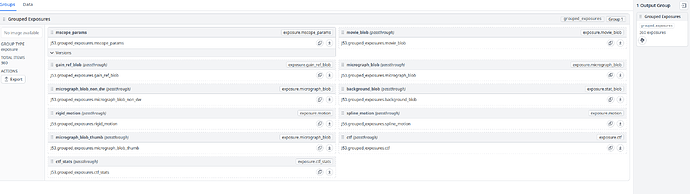
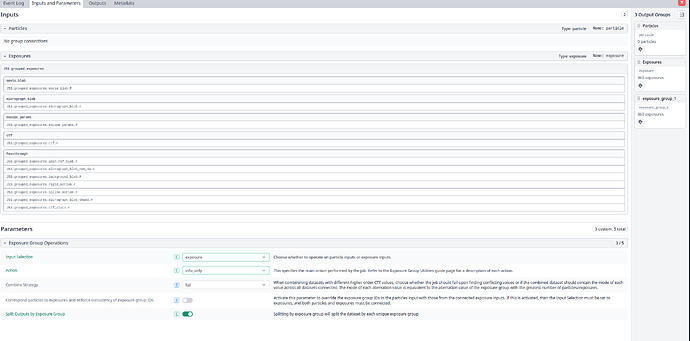
And here is my “Exposure Group Utilities” job:
Am I missing some other parameter that needs to be set?
EDIT: I just saw that there’s actually support for clustering right in CryoSPARC now, so I updated the beam_shift and beam_shift_known params and with the ImageShift values in the .mdoc files, and it’s working great now. I’m still curious why it didn’t find my manual groups, though.