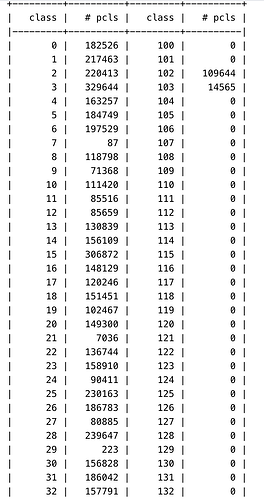
Running a 3D classification job with 7M particles and 200 classes, it runs fine for the O-EM iterations, but dies in the first full iteration with the attached error. Only other unusual thing is that a number of the classes at that point seem to be empty… I am running with less classes to see if that helps, but any thoughts as to the cause?
Outputting data...
Traceback (most recent call last):
File "cryosparc_master/cryosparc_compute/run.py", line 95, in cryosparc_master.cryosparc_compute.run.main
File "cryosparc_master/cryosparc_compute/jobs/class3D/run.py", line 951, in cryosparc_master.cryosparc_compute.jobs.class3D.run.run_class_3D
File "/home/exx/cryosparc/cryosparc_worker/cryosparc_tools/cryosparc/dataset.py", line 638, in save
outdata = self.to_records(fixed=True)
File "/home/exx/cryosparc/cryosparc_worker/cryosparc_tools/cryosparc/dataset.py", line 1267, in to_records
return numpy.core.records.fromarrays(arrays, dtype=dtype)
File "/home/exx/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.8/site-packages/numpy/core/records.py", line 643, in fromarrays
shape = arrayList[0].shape
IndexError: list index out of range
EDIT: I’m wondering if the completely empty classes might be causing the issue?
Hey @olibclarke – this is an error we’ve never encountered. No luck reproducing it internally so far. Could you tell us any non-default params for this job and paste the event log line (near the beginning of the job) that starts with All size parameters: {'N_p': ....}.
Many thanks!
Sure!
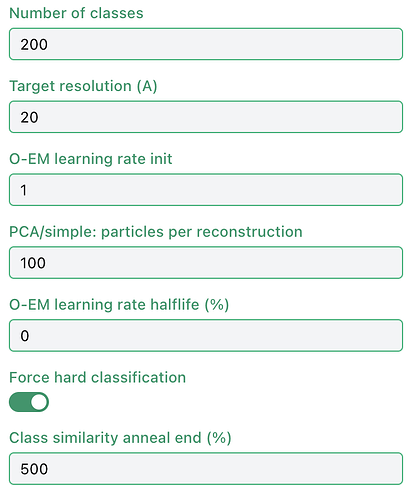
Custom params in screenshot (I realize the class similarity annealing has no effect when force hard classification is on, it was cloned from another job).
Here is the event log line requested:
All size parameters: {'N_p': 80, 'N_v': 80, 'N_cmp': 80, 'N_bp': 80, 'N_p_bp': 80, 'N_v_zp': 80, 'N_p_zp': 80, 'radwn_bp': 38, 'radwn_bp_v': 37, 'psize_v': 9.823049545288086, 'psize_p': 9.82305}
Thanks! Nothing of note… Could you also please check the job log output (under the Metadata tab → Log) for any errors related to the particle dataset?
Here are the last lines:
========= sending heartbeat at 2024-02-21 04:05:41.302192
out of memory (errno 1: Operation not permitted)
dset_ncol: invalid handle 18446744073709551615, no such slot (errno 1: Operation not permitted)
dset_ncol: invalid handle 18446744073709551615, no such slot (errno 1: Operation not permitted)
dset_ncol: invalid handle 18446744073709551615, no such slot (errno 1: Operation not permitted)
========= sending heartbeat at 2024-02-21 04:05:51.320091
HOST ALLOCATION FUNCTION: using numba.cuda.pinned_array
**** handle exception rc
/home/exx/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.8/multiprocessing/process.py:108: RuntimeWarning: divide by zero encountered in log
self._target(*self._args, **self._kwargs)
/home/exx/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.8/multiprocessing/process.py:108: RuntimeWarning: divide by zero encountered in log
self._target(*self._args, **self._kwargs)
/home/exx/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.8/multiprocessing/process.py:108: RuntimeWarning: divide by zero encountered in log
self._target(*self._args, **self._kwargs)
/home/exx/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.8/multiprocessing/process.py:108: RuntimeWarning: divide by zero encountered in true_divide
self._target(*self._args, **self._kwargs)
/home/exx/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.8/multiprocessing/process.py:108: RuntimeWarning: invalid value encountered in multiply
self._target(*self._args, **self._kwargs)
/home/exx/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.8/multiprocessing/process.py:108: RuntimeWarning: divide by zero encountered in log
self._target(*self._args, **self._kwargs)
/home/exx/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.8/multiprocessing/process.py:108: RuntimeWarning: invalid value encountered in true_divide
self._target(*self._args, **self._kwargs)
/home/exx/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.8/multiprocessing/process.py:108: RuntimeWarning: divide by zero encountered in true_divide
self._target(*self._args, **self._kwargs)
/home/exx/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.8/multiprocessing/process.py:108: RuntimeWarning: invalid value encountered in multiply
self._target(*self._args, **self._kwargs)
/home/exx/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.8/site-packages/numba/cuda/dispatcher.py:538: NumbaPerformanceWarning: Grid size 72 will likely result in GPU under-utilization due to low occupancy.
warn(NumbaPerformanceWarning(msg))
/home/exx/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.8/site-packages/numba/cuda/dispatcher.py:538: NumbaPerformanceWarning: Grid size 52 will likely result in GPU under-utilization due to low occupancy.
warn(NumbaPerformanceWarning(msg))
Traceback (most recent call last):
File "cryosparc_master/cryosparc_compute/run.py", line 95, in cryosparc_master.cryosparc_compute.run.main
File "cryosparc_master/cryosparc_compute/jobs/class3D/run.py", line 951, in cryosparc_master.cryosparc_compute.jobs.class3D.run.run_class_3D
File "/home/exx/cryosparc/cryosparc_worker/cryosparc_tools/cryosparc/dataset.py", line 638, in save
outdata = self.to_records(fixed=True)
File "/home/exx/cryosparc/cryosparc_worker/cryosparc_tools/cryosparc/dataset.py", line 1267, in to_records
return numpy.core.records.fromarrays(arrays, dtype=dtype)
File "/home/exx/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.8/site-packages/numpy/core/records.py", line 643, in fromarrays
shape = arrayList[0].shape
IndexError: list index out of range
set status to failed
========= main process now complete at 2024-02-21 04:05:56.434484.
========= monitor process now complete at 2024-02-21 04:05:56.441260.
Hi @olibclarke, based on the output log, it looks like the node ran out of CPU memory. We’ll make a fix to ensure this is more clear in a future release. As a workaround, you could try the classification at a lower resolution so that the job uses less memory.
1 Like
Thanks - indeed running with fewer classes it has got through a full iteration. It is already pretty low resolution though (20Å target res, particles downsampled to 80px box), and I have 256GB RAM on the workstation with nothing else running
1 Like