Hello.
Our researchers are having intermittent issues with 2D classification. Jobs are running in the pipeline, so there should not be any missing files, but we get the attached error. If we clear the job and re-run or lower the number of classes, if will then work.
What could be the root cause of this issue?
Hi @yodamoppet, thanks for posting. We are looking into this. Can you please copy-paste the text of the traceback for future searchability?
Excellent.
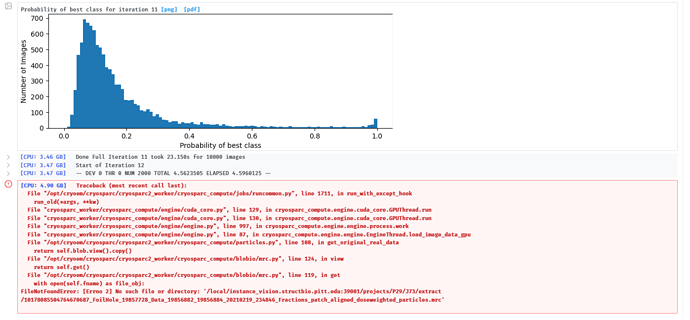
Certainly, here is the traceback text:
[CPU: 4.90 GB] Traceback (most recent call last):
File °/opt/cryoem/cryosparc/cryosparc2_worker/cryosparc_compute/jobs/runcommon.py”, line 1711, in run_with_except_hook
run_old(*args, **kw)
File “cryosparc_worker/cryosparc_compute/engine/cuda_core.py”, line 129, in cryosparc_compute.engine.cuda_core.GPUThread.run
File “cryosparc_worker/cryosparc_compute/engine/cuda_core.py”, line 136, in cryosparc_compute.engine.cuda_core.GPUThread.run
File “cryosparc_worker/cryosparc_compute/engine/engine.py”, line 997, in cryosparc_compute.engine.engine.process.work
File “cryosparc_worker/cryosparc_compute/engine/engine.py”, line 87, in cryosparc_compute.engine. engine. EngineThread. load_image_dat
File °/opt/cryoem/cryosparc/cryosparc2_worker/cryosparc_compute/particles.py”, line 168, in get_original_real_data
return self.blob.view().copy{)
File °/opt/cryoem/cryosparc/cryosparc2_worker/cryosparc_compute/blobio/mrc.py”, line 124, in view
return self.get(}
File °/opt/cryoem/cryosparc/cryosparc2_worker/cryosparc_compute/blobio/mrc.py”, line 119, in get
with open(self. fname) as file_obj:
FileNotFoundError: [Errno 2] Mo such file or directory: ‘/lLocal/instance_vision.structbio.pitt.edu:39061/projects/P29/J73/extract
/10178085504764670687_FoilHole_19857728_Data_19856882_19856884_20210219_234846_Fractions_patch_aligned_doseweighted_particles.mrc’
Hi @yodamoppet,
Sorry for the delay. This traceback looks like the actual file from the cache folder was deleted. Is it possible someone on this same machine deleted all the files on that volume to make space for their work? Or is it possible there is a script or task that runs and clears out the cache volume intermittently?
Hi Stephen,
Sorry for the delay in getting back to this.
Yes, after tracing, this appears to be precisely what has happened. The cache drive filled and was deleted by a script monitoring this situation. I have disabled it for now.
I notice that there is an option “–ssdquota” which will allow me to limit the quota space the worker uses. Is this something that can be added to config.sh for the worked (i.e. “export SSDQUOTA=500GB” or similar), or do I need to recompile the worker to get this functionality?
Thanks!
Hi @yodamoppet,
Yes, you can update a worker’s configuation easily by running the ./bin/cryosparcw connect command with the --update flag:
https://guide.cryosparc.com/setup-configuration-and-management/how-to-download-install-and-configure/downloading-and-installing-cryosparc#connect-the-standalone-worker-node-to-the-master-node
./bin/cryosparcw connect --worker <worker_hostname> \
--master <master_hostname> \
--port <port_num> \
--update \
--ssdquota <ssd_quota_mb>
So, in this case, using “–update” will set the ssdquota without messing with other parameters. For example:
./cryosparcw connect --update --ssdquota 1000000
to give the worker a quota of 1tb without adjusting any other previously configured parameters?
Thanks for the assistance!
You will need to provide the worker, master and port arguments as well (to indicate which worker you’d like to update)
Very good, thank you.
Is there any way to output/verify what was used for worker, master, port?
Can this be done while jobs are running, or will it interrupt jobs?
Hi @yodamoppet,
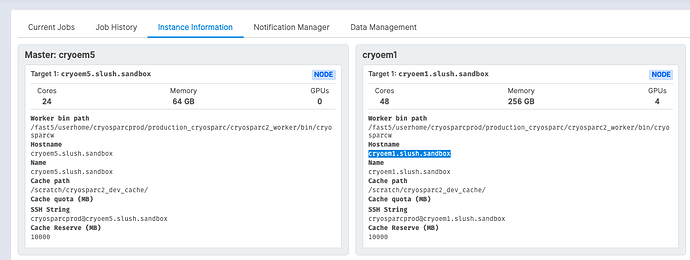
You can look at the “Instance Information” tab in the Resource Manager to see the argument for worker (Hostname):
The master and port arguments will be found in cryosparc_master/config.sh as CRYOSPARC_MASTER_HOSTNAME and CRYOSPARC_BASE_PORT respectively.
Yes, it won’t interrupt any running jobs.
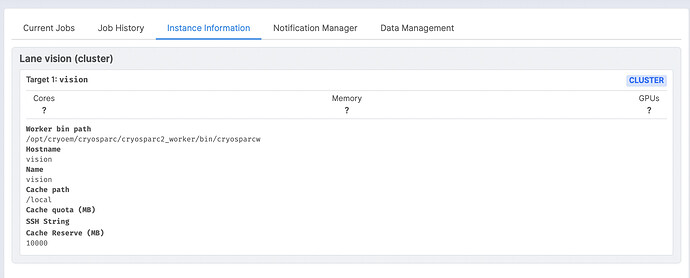
Thanks for the details. I’ve mostly got this ready to try, but I’m not sure what to use for the --worker argument. This is a cluster system by the way. I don’t see workers listed as in your “Instance Information” tab (see image).
What in this case is the appropriate argument to use for “–worker” in the update command?
Hey @yodamoppet,
In the case of a cluster, the way to update the configuration is different.
https://guide.cryosparc.com/setup-configuration-and-management/how-to-download-install-and-configure/downloading-and-installing-cryosparc#update-a-cluster-configuration
To update an existing cluster integration, call the cryosparcm cluster connect commands with the updated cluster_info.json and cluster_script.sh in the current working directory.
Note that the name field from cluster_info.json must be the same in the cluster configuration you’re trying to update
If you don’t already have the cluster_info.json and cluster_script.sh files in your current working directory, you can get them by running the command cryosparcm cluster dump <name>
1 Like
Hello.
Thanks for the clarification. I have the cluster_info.json and cluster_script.sh files in the cryosparcm directory.
What arguments do I need to add to these files to limit the ssd quota? It looks like this would mainly go in cluster_info.json, right?
Current cluster_info.json is:
{
“name” : “vision”,
“worker_bin_path” : “/opt/cryoem/cryosparc/cryosparc2_worker/bin/cryosparcw”,
“cache_path” : “/local”,
“send_cmd_tpl” : “{{ command }}”,
“qsub_cmd_tpl” : “sbatch {{ script_path_abs }}”,
“qstat_cmd_tpl” : “squeue -j {{ cluster_job_id }}”,
“qdel_cmd_tpl” : “scancel {{ cluster_job_id }}”,
“qinfo_cmd_tpl” : “sinfo”,
“transfer_cmd_tpl” : “scp {{ src_path }} loginnode:{{ dest_path }}”
}
Hi @yodamoppet,
Correct, in the cluster_info.json file, you can add a new field cache_quota_mb, with the value in MB.
For example, if you wanted a 1TB quota, your file will look like:
{
“name” : “vision”,
“worker_bin_path” : “/opt/cryoem/cryosparc/cryosparc2_worker/bin/cryosparcw”,
“cache_path” : “/local”,
“send_cmd_tpl” : “{{ command }}”,
“qsub_cmd_tpl” : “sbatch {{ script_path_abs }}”,
“qstat_cmd_tpl” : “squeue -j {{ cluster_job_id }}”,
“qdel_cmd_tpl” : “scancel {{ cluster_job_id }}”,
“qinfo_cmd_tpl” : “sinfo”,
“transfer_cmd_tpl” : “scp {{ src_path }} loginnode:{{ dest_path }}”,
"cache_quota_mb" : 1000000
}
Very good. Two additional questions:
As far as syntax, should there be a closing quote after the 1000000 ?
And, will this interrupt jobs, and do I need to stop or restart the cryoapracm process at any stage?
Hi @yodamoppet,
The cryoSPARC function that reads this JSON file expects a number for this field, so quotes aren’t required.
View the fields info here, including their datatype:
https://guide.cryosparc.com/setup-configuration-and-management/how-to-download-install-and-configure/downloading-and-installing-cryosparc#create-the-files
This will not interrupt any currently running jobs. You can safely run this command while cryoSPARC is running and while jobs are running, no need to restart.
Very good, I have just executed this.
I get the a message after executing:
Successfully added cluster lane vision
Is that the correct completion message? I expected something like “successfully updated cluster lane vision”, but perhaps the message is the same whether adding a lane or updating it?