Hi All,
I am trying to solve the structure of a Fab-antigen complex. It has a relatively complex quaternary structure (two chains in Fab, three chains in antigen), and the particles are not particularly large (only 50 and 130 Angstrom diameters at minimum and maximum, respectively). We know the structures of the antigen and of the Fab independently, but would like to characterize the structure of the complex, to ascertain the Fab epitope.
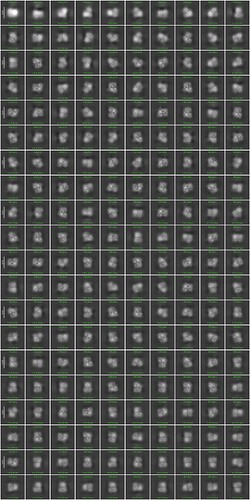
After pre-processing (motion correction + CTF estimation) and curation of exposures, 2,008 exposures were used for (1) manual picking followed by (2) template picking. Template picks were then used to train a Topaz model. Topaz selected 193,353 particles; however, 2D classes have a pixelated appearance (see image below), and Ab Initio models (4 classes) are not producing a structure that appears close to what I expect.
How could I improve my output? Thank you all for your time and guidance.
Best Regards,