Dear All,
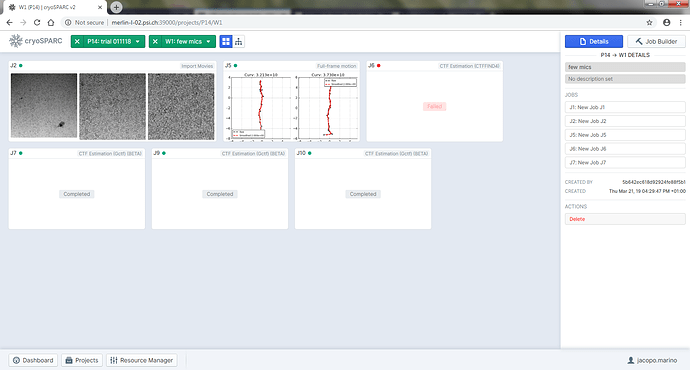
sorry to keep asking about this issue, but I find interesting that also GCTF fails, despite the fact that no error is reported. Here is the output:
Importing job module for job type ctf_estimation_gctf…
Job ready to run
Wrap of Gctf_v1.06
Zhang, K. (2016). Gctf : Real-time CTF determination and correction. Journal of Structural Biology, 193(1), 1-12. https://doi.org/10.1016/j.jsb.2015.11.003
Using files in wildcard path /gpfs/group/LBR/p007/Jacopo/011118/part1/P14/J10/motioncorrected/FoilHole_9180*.mrc
Writing /gpfs/group/LBR/p007/Jacopo/011118/part1/P14/J10/input_ctfstar.star
Gctf command: /gpfs/data/marino_j/cryosparc/v2/cryosparc/cryosparc2_worker/deps/external/gctf-1.06/bin/Gctf-v1.06_sm_20_cu5.0_x86_64 --apix 0.854 --kV 300.0 --Cs 2.7 --ac 0.1 --ctfstar /gpfs/group/LBR/p007/Jacopo/011118/part1/P14/J10/micrographs_all_gctf.star --input_ctfstar /gpfs/group/LBR/p007/Jacopo/011118/part1/P14/J10/input_ctfstar.star --gid 0 --boxsize 1024 --boxsuffix _automatch.star --logsuffix _gctf.log
ERROR [Errno 2] No such file or directory
exposure_blobpath_to_dsetidx_d {u’J10/motioncorrected/FoilHole_918071_Data_922094_922095_20181101_1715-8456_rigid_aligned.mrc’: 42, u’J10/motioncorrected/FoilHole_918063_Data_922111_922112_20181101_1648-8420_rigid_aligned.mrc’: 6, u’J10/motioncorrected/FoilHole_918080_Data_922111_922112_20181101_1743-8498_rigid_aligned.mrc’: 84, u’J10/motioncorrected/FoilHole_918084_Data_922122_922123_20181101_1755-8515_rigid_aligned.mrc’: 105, u’J10/motioncorrected/FoilHole_918070_Data_921860_921861_20181101_1711-8449_rigid_aligned.mrc’: 36, u’J10/motioncorrected/FoilHole_918065_Data_922111_922112_20181101_1655-8430_rigid_aligned.mrc’: 16, u’J10/motioncorrected/FoilHole_918068_Data_921860_921861_20181101_1705-8439_rigid_aligned.mrc’: 26, u’J10/motioncorrected/FoilHole_918079_Data_922111_922112_20181101_1740-8493_rigid_aligned.mrc’: 79, u’J10/motioncorrected/FoilHole_918062_Data_922122_922123_20181101_1601-8413_rigid_aligned.mrc’: 2, u’J10/motioncorrected/FoilHole_918081_Data_922122_922123_20181101_1745-8500_rigid_aligned.mrc’: 90, u’J10/motioncorrected/FoilHole_918075_Data_922122_922123_20181101_1728-8475_rigid_aligned.mrc’: 65, u’J10/motioncorrected/FoilHole_918074_Data_922094_922095_20181101_1725-8471_rigid_aligned.mrc’: 57, u’J10/motioncorrected/FoilHole_918078_Data_922111_922112_20181101_1737-8488_rigid_aligned.mrc’: 74, u’J10/motioncorrected/FoilHole_918065_Data_922102_922103_20181101_1655-8429_rigid_aligned.mrc’: 15, u’J10/motioncorrected/FoilHole_918073_Data_922102_922103_20181101_1722-8467_rigid_aligned.mrc’: 53, u’J10/motioncorrected/FoilHole_918069_Data_922111_922112_20181101_1710-8448_rigid_aligned.mrc’: 34, u’J10/motioncorrected/FoilHole_918072_Data_922111_922112_20181101_1720-8463_rigid_aligned.mrc’: 49, u’J10/motioncorrected/FoilHole_918078_Data_922102_922103_20181101_1737-8487_rigid_aligned.mrc’: 73, u’J10/motioncorrected/FoilHole_918071_Data_922102_922103_20181101_1716-8457_rigid_aligned.mrc’: 43, u’J10/motioncorrected/FoilHole_918083_Data_922111_922112_20181101_1753-8513_rigid_aligned.mrc’: 99, u’J10/motioncorrected/FoilHole_918065_Data_922122_922123_20181101_1654-8427_rigid_aligned.mrc’: 17, u’J10/motioncorrected/FoilHole_918062_Data_921860_921861_20181101_1601-8412_rigid_aligned.mrc’: 0, u’J10/motioncorrected/FoilHole_918083_Data_921860_921861_20181101_1751-8509_rigid_aligned.mrc’: 96, u’J10/motioncorrected/FoilHole_918073_Data_922111_922112_20181101_1723-8468_rigid_aligned.mrc’: 54, u’J10/motioncorrected/FoilHole_918068_Data_922111_922112_20181101_1706-8443_rigid_aligned.mrc’: 29, u’J10/motioncorrected/FoilHole_918063_Data_921860_921861_20181101_1647-8416_rigid_aligned.mrc’: 3, u’J10/motioncorrected/FoilHole_918079_Data_921860_921861_20181101_1739-8489_rigid_aligned.mrc’: 76, u’J10/motioncorrected/FoilHole_918082_Data_922094_922095_20181101_1749-8506_rigid_aligned.mrc’: 92, u’J10/motioncorrected/FoilHole_918070_Data_922111_922112_20181101_1713-8453_rigid_aligned.mrc’: 39, u’J10/motioncorrected/FoilHole_918072_Data_921860_921861_20181101_1718-8459_rigid_aligned.mrc’: 46, u’J10/motioncorrected/FoilHole_918079_Data_922122_922123_20181101_1739-8490_rigid_aligned.mrc’: 80, u’J10/motioncorrected/FoilHole_918071_Data_922122_922123_20181101_1715-8455_rigid_aligned.mrc’: 45, u’J10/motioncorrected/FoilHole_918079_Data_922102_922103_20181101_1740-8492_rigid_aligned.mrc’: 78, u’J10/motioncorrected/FoilHole_918069_Data_922102_922103_20181101_1709-8447_rigid_aligned.mrc’: 33, u’J10/motioncorrected/FoilHole_918070_Data_922094_922095_20181101_1712-8451_rigid_aligned.mrc’: 37, u’J10/motioncorrected/FoilHole_918070_Data_922122_922123_20181101_1712-8450_rigid_aligned.mrc’: 40, u’J10/motioncorrected/FoilHole_918067_Data_922094_922095_20181101_1702-8436_rigid_aligned.mrc’: 22, u’J10/motioncorrected/FoilHole_918080_Data_922122_922123_20181101_1742-8495_rigid_aligned.mrc’: 85, u’J10/motioncorrected/FoilHole_918068_Data_922122_922123_20181101_1705-8440_rigid_aligned.mrc’: 30, u’J10/motioncorrected/FoilHole_918067_Data_922122_922123_20181101_1702-8435_rigid_aligned.mrc’: 25, u’J10/motioncorrected/FoilHole_918082_Data_921860_921861_20181101_1748-8504_rigid_aligned.mrc’: 91, u’J10/motioncorrected/FoilHole_918072_Data_922122_922123_20181101_1719-8460_rigid_aligned.mrc’: 50, u’J10/motioncorrected/FoilHole_918074_Data_922122_922123_20181101_1725-8470_rigid_aligned.mrc’: 60, u’J10/motioncorrected/FoilHole_918066_Data_922094_922095_20181101_1658-8433_rigid_aligned.mrc’: 19, u’J10/motioncorrected/FoilHole_918078_Data_922094_922095_20181101_1736-8486_rigid_aligned.mrc’: 72, u’J10/motioncorrected/FoilHole_918062_Data_922094_922095_20181101_1601-8414_rigid_aligned.mrc’: 1, u’J10/motioncorrected/FoilHole_918073_Data_921860_921861_20181101_1721-8464_rigid_aligned.mrc’: 51, u’J10/motioncorrected/FoilHole_918068_Data_922102_922103_20181101_1706-8442_rigid_aligned.mrc’: 28, u’J10/motioncorrected/FoilHole_918075_Data_922094_922095_20181101_1729-8476_rigid_aligned.mrc’: 62, u’J10/motioncorrected/FoilHole_918085_Data_921860_921861_20181101_1757-8519_rigid_aligned.mrc’: 106, u’J10/motioncorrected/FoilHole_918084_Data_922111_922112_20181101_1756-8518_rigid_aligned.mrc’: 104, u’J10/motioncorrected/FoilHole_918073_Data_922122_922123_20181101_1722-8465_rigid_aligned.mrc’: 55, u’J10/motioncorrected/FoilHole_918083_Data_922094_922095_20181101_1752-8511_rigid_aligned.mrc’: 97, u’J10/motioncorrected/FoilHole_918075_Data_922102_922103_20181101_1729-8477_rigid_aligned.mrc’: 63, u’J10/motioncorrected/FoilHole_918068_Data_922094_922095_20181101_1705-8441_rigid_aligned.mrc’: 27, u’J10/motioncorrected/FoilHole_918081_Data_921860_921861_20181101_1745-8499_rigid_aligned.mrc’: 86, u’J10/motioncorrected/FoilHole_918082_Data_922102_922103_20181101_1749-8507_rigid_aligned.mrc’: 93, u’J10/motioncorrected/FoilHole_918085_Data_922094_922095_20181101_1758-8521_rigid_aligned.mrc’: 107, u’J10/motioncorrected/FoilHole_918064_Data_922102_922103_20181101_1651-8424_rigid_aligned.mrc’: 10, u’J10/motioncorrected/FoilHole_918076_Data_921860_921861_20181101_1732-8479_rigid_aligned.mrc’: 66, u’J10/motioncorrected/FoilHole_918064_Data_921860_921861_20181101_1650-8421_rigid_aligned.mrc’: 8, u’J10/motioncorrected/FoilHole_918071_Data_921860_921861_20181101_1715-8454_rigid_aligned.mrc’: 41, u’J10/motioncorrected/FoilHole_918066_Data_922122_922123_20181101_1658-8432_rigid_aligned.mrc’: 20, u’J10/motioncorrected/FoilHole_918072_Data_922094_922095_20181101_1719-8461_rigid_aligned.mrc’: 47, u’J10/motioncorrected/FoilHole_918082_Data_922111_922112_20181101_1749-8508_rigid_aligned.mrc’: 94, u’J10/motioncorrected/FoilHole_918075_Data_922111_922112_20181101_1730-8478_rigid_aligned.mrc’: 64, u’J10/motioncorrected/FoilHole_918067_Data_922111_922112_20181101_1703-8438_rigid_aligned.mrc’: 24, u’J10/motioncorrected/FoilHole_918067_Data_921860_921861_20181101_1701-8434_rigid_aligned.mrc’: 21, u’J10/motioncorrected/FoilHole_918063_Data_922102_922103_20181101_1648-8419_rigid_aligned.mrc’: 5, u’J10/motioncorrected/FoilHole_918080_Data_922102_922103_20181101_1743-8497_rigid_aligned.mrc’: 83, u’J10/motioncorrected/FoilHole_918082_Data_922122_922123_20181101_1748-8505_rigid_aligned.mrc’: 95, u’J10/motioncorrected/FoilHole_918074_Data_922102_922103_20181101_1726-8472_rigid_aligned.mrc’: 58, u’J10/motioncorrected/FoilHole_918073_Data_922094_922095_20181101_1722-8466_rigid_aligned.mrc’: 52, u’J10/motioncorrected/FoilHole_918065_Data_922094_922095_20181101_1654-8428_rigid_aligned.mrc’: 14, u’J10/motioncorrected/FoilHole_918074_Data_922111_922112_20181101_1726-8473_rigid_aligned.mrc’: 59, u’J10/motioncorrected/FoilHole_918072_Data_922102_922103_20181101_1719-8462_rigid_aligned.mrc’: 48, u’J10/motioncorrected/FoilHole_918064_Data_922094_922095_20181101_1651-8423_rigid_aligned.mrc’: 9, u’J10/motioncorrected/FoilHole_918076_Data_922111_922112_20181101_1733-8483_rigid_aligned.mrc’: 69, u’J10/motioncorrected/FoilHole_918069_Data_922094_922095_20181101_1709-8446_rigid_aligned.mrc’: 32, u’J10/motioncorrected/FoilHole_918083_Data_922102_922103_20181101_1752-8512_rigid_aligned.mrc’: 98, u’J10/motioncorrected/FoilHole_918081_Data_922111_922112_20181101_1746-8503_rigid_aligned.mrc’: 89, u’J10/motioncorrected/FoilHole_918074_Data_921860_921861_20181101_1725-8469_rigid_aligned.mrc’: 56, u’J10/motioncorrected/FoilHole_918084_Data_922094_922095_20181101_1755-8516_rigid_aligned.mrc’: 102, u’J10/motioncorrected/FoilHole_918079_Data_922094_922095_20181101_1739-8491_rigid_aligned.mrc’: 77, u’J10/motioncorrected/FoilHole_918064_Data_922122_922123_20181101_1650-8422_rigid_aligned.mrc’: 12, u’J10/motioncorrected/FoilHole_918075_Data_921860_921861_20181101_1728-8474_rigid_aligned.mrc’: 61, u’J10/motioncorrected/FoilHole_918064_Data_922111_922112_20181101_1652-8425_rigid_aligned.mrc’: 11, u’J10/motioncorrected/FoilHole_918067_Data_922102_922103_20181101_1702-8437_rigid_aligned.mrc’: 23, u’J10/motioncorrected/FoilHole_918078_Data_921860_921861_20181101_1736-8484_rigid_aligned.mrc’: 71, u’J10/motioncorrected/FoilHole_918081_Data_922094_922095_20181101_1746-8501_rigid_aligned.mrc’: 87, u’J10/motioncorrected/FoilHole_918078_Data_922122_922123_20181101_1736-8485_rigid_aligned.mrc’: 75, u’J10/motioncorrected/FoilHole_918084_Data_922102_922103_20181101_1755-8517_rigid_aligned.mrc’: 103, u’J10/motioncorrected/FoilHole_918066_Data_921860_921861_20181101_1657-8431_rigid_aligned.mrc’: 18, u’J10/motioncorrected/FoilHole_918063_Data_922122_922123_20181101_1647-8417_rigid_aligned.mrc’: 7, u’J10/motioncorrected/FoilHole_918069_Data_922122_922123_20181101_1709-8445_rigid_aligned.mrc’: 35, u’J10/motioncorrected/FoilHole_918085_Data_922102_922103_20181101_1758-8522_rigid_aligned.mrc’: 108, u’J10/motioncorrected/FoilHole_918084_Data_921860_921861_20181101_1754-8514_rigid_aligned.mrc’: 101, u’J10/motioncorrected/FoilHole_918085_Data_922111_922112_20181101_1759-8523_rigid_aligned.mrc’: 109, u’J10/motioncorrected/FoilHole_918085_Data_922122_922123_20181101_1758-8520_rigid_aligned.mrc’: 110, u’J10/motioncorrected/FoilHole_918071_Data_922111_922112_20181101_1716-8458_rigid_aligned.mrc’: 44, u’J10/motioncorrected/FoilHole_918080_Data_921860_921861_20181101_1742-8494_rigid_aligned.mrc’: 81, u’J10/motioncorrected/FoilHole_918063_Data_922094_922095_20181101_1648-8418_rigid_aligned.mrc’: 4, u’J10/motioncorrected/FoilHole_918076_Data_922094_922095_20181101_1732-8481_rigid_aligned.mrc’: 67, u’J10/motioncorrected/FoilHole_918080_Data_922094_922095_20181101_1743-8496_rigid_aligned.mrc’: 82, u’J10/motioncorrected/FoilHole_918081_Data_922102_922103_20181101_1746-8502_rigid_aligned.mrc’: 88, u’J10/motioncorrected/FoilHole_918069_Data_921860_921861_20181101_1708-8444_rigid_aligned.mrc’: 31, u’J10/motioncorrected/FoilHole_918070_Data_922102_922103_20181101_1713-8452_rigid_aligned.mrc’: 38, u’J10/motioncorrected/FoilHole_918076_Data_922122_922123_20181101_1732-8480_rigid_aligned.mrc’: 70, u’J10/motioncorrected/FoilHole_918065_Data_921860_921861_20181101_1654-8426_rigid_aligned.mrc’: 13, u’J10/motioncorrected/FoilHole_918076_Data_922102_922103_20181101_1733-8482_rigid_aligned.mrc’: 68, u’J10/motioncorrected/FoilHole_918083_Data_922122_922123_20181101_1752-8510_rigid_aligned.mrc’: 100}
initial fail_idx_set set([0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110])
ERROR: Gctf failed: [’/gpfs/data/marino_j/cryosparc/v2/cryosparc/cryosparc2_worker/deps/external/gctf-1.06/bin/Gctf-v1.06_sm_20_cu5.0_x86_64’, ‘–apix’, ‘0.854’, ‘–kV’, ‘300.0’, ‘–Cs’, ‘2.7’, ‘–ac’, ‘0.1’, ‘–ctfstar’, ‘/gpfs/group/LBR/p007/Jacopo/011118/part1/P14/J10/micrographs_all_gctf.star’, ‘–input_ctfstar’, ‘/gpfs/group/LBR/p007/Jacopo/011118/part1/P14/J10/input_ctfstar.star’, ‘–gid’, ‘0’, ‘–boxsize’, ‘1024’, ‘–boxsuffix’, ‘_automatch.star’, ‘–logsuffix’, ‘_gctf.log’]
ERROR: 111 exposures failed: [’/gpfs/data/marino_j/cryosparc/v2/cryosparc/cryosparc2_worker/deps/external/gctf-1.06/bin/Gctf-v1.06_sm_20_cu5.0_x86_64’, ‘–apix’, ‘0.854’, ‘–kV’, ‘300.0’, ‘–Cs’, ‘2.7’, ‘–ac’, ‘0.1’, ‘–ctfstar’, ‘/gpfs/group/LBR/p007/Jacopo/011118/part1/P14/J10/micrographs_all_gctf.star’, ‘–input_ctfstar’, ‘/gpfs/group/LBR/p007/Jacopo/011118/part1/P14/J10/input_ctfstar.star’, ‘–gid’, ‘0’, ‘–boxsize’, ‘1024’, ‘–boxsuffix’, ‘_automatch.star’, ‘–logsuffix’, ‘_gctf.log’]
Completed CTF Estimation using Gctf
Zhang, K. (2016). Gctf : Real-time CTF determination and correction. Journal of Structural Biology, 193(1), 1-12. https://doi.org/10.1016/j.jsb.2015.11.003
Compiling job outputs…
Passing through outputs for output group exposures_success from input group exposures
This job outputted results [‘ctf’, ‘ctf_plotdata’]
Loaded output dset with 0 items
Passthrough results [‘movie_blob’, ‘micrograph_blob_non_dw’, ‘micrograph_blob’, ‘mscope_params’, ‘background_blob’, ‘rigid_motion’, ‘gain_ref_blob’]
Loaded passthrough dset with 111 items
Intersection of output and passthrough has 0 items
Passing through outputs for output group exposures_fail from input group exposures
This job outputted results [‘ctf’, ‘ctf_plotdata’]
Loaded output dset with 111 items
Passthrough results [‘movie_blob’, ‘micrograph_blob_non_dw’, ‘micrograph_blob’, ‘mscope_params’, ‘background_blob’, ‘rigid_motion’, ‘gain_ref_blob’]
Loaded passthrough dset with 111 items
Intersection of output and passthrough has 111 items
Passing through outputs for output group particles_success from input group particles
This job outputted results [‘ctf’]
Loaded output dset with 0 items
Passthrough results []
Passing through outputs for output group particles_fail from input group particles
This job outputted results [‘ctf’]
Loaded output dset with 0 items
Passthrough results []
Checking outputs for output group exposures_success
Checking outputs for output group exposures_fail
Checking outputs for output group particles_success
Checking outputs for output group particles_fail
Job complete. Total time 0.68s