Here you go:
Launching job on lane default target ubuntu ...
Running job on master node hostname ubuntu
[CPU: 105.4 MB] Project P22 Job J14 Started
[CPU: 105.5 MB] Master running v2.16.1-live_deeppick_privatebeta+200722, worker running v2.16.1-live_deeppick_privatebeta+200722
[CPU: 105.6 MB] Running on lane default
[CPU: 105.6 MB] Resources allocated:
[CPU: 105.9 MB] Worker: ubuntu
[CPU: 105.9 MB] CPU : [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23]
[CPU: 105.9 MB] GPU : [0]
[CPU: 105.9 MB] RAM : [0]
[CPU: 105.9 MB] SSD : False
[CPU: 105.9 MB] --------------------------------------------------------------
[CPU: 105.9 MB] Importing job module for job type topaz_train...
[CPU: 215.3 MB] Job ready to run
[CPU: 215.3 MB] ***************************************************************
[CPU: 215.3 MB] Topaz is a particle detection tool created by Tristan Bepler and Alex J. Noble.
Citations:
- Bepler, T., Morin, A., Rapp, M. et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat Methods 16, 1153-1160 (2019) doi:10.1038/s41592-019-0575-8
- Bepler, T., Noble, A.J., Berger, B. Topaz-Denoise: general deep denoising models for cryoEM. bioRxiv 838920 (2019) doi: https://doi.org/10.1101/838920
Structura Biotechnology Inc. and cryoSPARC do not license Topaz nor distribute Topaz binaries. Please ensure you have your own copy of Topaz licensed and installed under the terms of its GNU General Public License v3.0, available for review at: https://github.com/tbepler/topaz/blob/master/LICENSE.
***************************************************************
[CPU: 216.0 MB] Starting Topaz process using version 0.2.4...
[CPU: 216.0 MB] Random seed used is 1979721300
[CPU: 216.0 MB] --------------------------------------------------------------
[CPU: 216.0 MB] Starting preprocessing...
[CPU: 216.0 MB] Starting micrograph preprocessing by running command /home/user/software/anaconda3/envs/topaz/bin/topaz preprocess --scale 4 --niters 200 --num-workers 24 -o /home/user/processing/cryosparc_projects/empiar/S_protein/P22/J14/preprocessed [MICROGRAPH PATHS EXCLUDED FOR LEGIBILITY]
[CPU: 216.0 MB] Preprocessing over 8 processes...
[CPU: 216.1 MB] Inverting negative staining...
[CPU: 216.1 MB] Inverting negative staining complete.
[CPU: 216.1 MB] Micrograph preprocessing command complete.
[CPU: 216.1 MB] Starting particle pick preprocessing by running command /home/user/software/anaconda3/envs/topaz/bin/topaz convert --down-scale 4 --threshold 0 -o /home/user/processing/cryosparc_projects/empiar/S_protein/P22/J14/topaz_particles_processed.txt /home/user/processing/cryosparc_projects/empiar/S_protein/P22/J14/topaz_particles_raw.txt
[CPU: 216.1 MB] Particle pick preprocessing command complete.
[CPU: 216.1 MB] Preprocessing done in 360.565s.
[CPU: 216.1 MB] --------------------------------------------------------------
[CPU: 216.1 MB] Starting train-test splitting...
[CPU: 216.1 MB] Starting dataset splitting by running command /home/user/software/anaconda3/envs/topaz/bin/topaz train_test_split --number 20 --seed 1979721300 --image-dir /home/user/processing/cryosparc_projects/empiar/S_protein/P22/J14/preprocessed /home/user/processing/cryosparc_projects/empiar/S_protein/P22/J14/topaz_particles_processed.txt
[CPU: 216.1 MB] # splitting 100 micrographs with 2242 labeled particles into 80 train and 20 test micrographs
[CPU: 216.1 MB] WARNING: no micrograph found matching image name "n20apr21a_b2g2_00010gr_00017sq_v02_00002hln_00007enn-a-DW". Skipping it.
[CPU: 216.1 MB] WARNING: no micrograph found matching image name "n20apr21a_b2g2_00016gr_00053sq_v02_00002hln_00011enn-a-DW". Skipping it.
[CPU: 216.1 MB] WARNING: no micrograph found matching image name "n20apr21a_b3g1_00022gr_00052sq_v02_00002hln_v01_00005enn-a-DW". Skipping it.
[CPU: 216.1 MB] WARNING: no micrograph found matching image name "n20apr21a_b2g2_00015gr_00012sq_v02_00004hln_00002enn-a-DW". Skipping it.
[CPU: 216.1 MB] WARNING: no micrograph found matching image name "n20apr21a_b2g2_00015gr_00007sq_v02_00002hln_00002enn-a-DW". Skipping it.
[CPU: 216.1 MB] WARNING: no micrograph found matching image name "n20apr21a_b2g2_00016gr_00082sq_v02_00002hln_v01_00011enn-a-DW". Skipping it.
[CPU: 216.1 MB] WARNING: no micrograph found matching image name "n20apr21a_b2g2_00015gr_00012sq_v02_00004hln_00003enn-a-DW". Skipping it.
[CPU: 216.1 MB] Traceback (most recent call last):
[CPU: 216.1 MB] File "/home/user/software/anaconda3/envs/topaz/bin/topaz", line 11, in <module>
[CPU: 216.1 MB] load_entry_point('topaz-em==0.2.4', 'console_scripts', 'topaz')()
[CPU: 216.1 MB] File "/home/user/software/anaconda3/envs/topaz/lib/python3.6/site-packages/topaz/main.py", line 148, in main
[CPU: 216.1 MB] args.func(args)
[CPU: 216.1 MB] File "/home/user/software/anaconda3/envs/topaz/lib/python3.6/site-packages/topaz/commands/train_test_split.py", line 128, in main
[CPU: 216.1 MB] image_list_train = pd.DataFrame({'image_name': image_names_train, 'path': paths_train})
[CPU: 216.1 MB] File "/home/user/software/anaconda3/envs/topaz/lib/python3.6/site-packages/pandas/core/frame.py", line 435, in __init__
[CPU: 216.1 MB] mgr = init_dict(data, index, columns, dtype=dtype)
[CPU: 216.1 MB] File "/home/user/software/anaconda3/envs/topaz/lib/python3.6/site-packages/pandas/core/internals/construction.py", line 254, in init_dict
[CPU: 216.1 MB] return arrays_to_mgr(arrays, data_names, index, columns, dtype=dtype)
[CPU: 216.1 MB] File "/home/user/software/anaconda3/envs/topaz/lib/python3.6/site-packages/pandas/core/internals/construction.py", line 64, in arrays_to_mgr
[CPU: 216.1 MB] index = extract_index(arrays)
[CPU: 216.1 MB] File "/home/user/software/anaconda3/envs/topaz/lib/python3.6/site-packages/pandas/core/internals/construction.py", line 365, in extract_index
[CPU: 216.1 MB] raise ValueError("arrays must all be same length")
[CPU: 216.1 MB] ValueError: arrays must all be same length
[CPU: 216.1 MB]
Dataset splitting command complete.
[CPU: 216.1 MB] Train-test splitting done in 1.658s.
[CPU: 216.1 MB] --------------------------------------------------------------
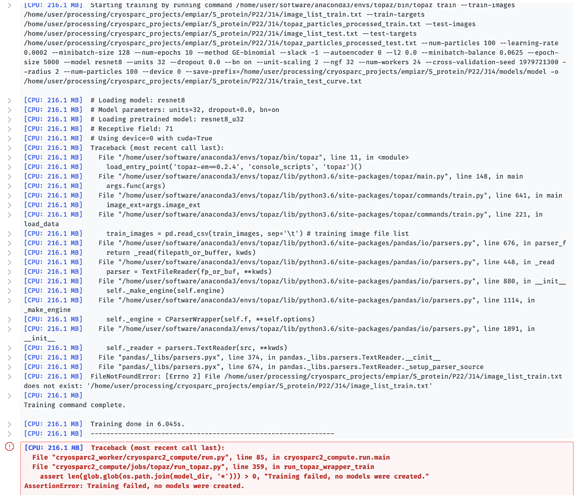
[CPU: 216.1 MB] Starting training...
[CPU: 216.1 MB] Starting training by running command /home/user/software/anaconda3/envs/topaz/bin/topaz train --train-images /home/user/processing/cryosparc_projects/empiar/S_protein/P22/J14/image_list_train.txt --train-targets /home/user/processing/cryosparc_projects/empiar/S_protein/P22/J14/topaz_particles_processed_train.txt --test-images /home/user/processing/cryosparc_projects/empiar/S_protein/P22/J14/image_list_test.txt --test-targets /home/user/processing/cryosparc_projects/empiar/S_protein/P22/J14/topaz_particles_processed_test.txt --num-particles 100 --learning-rate 0.0002 --minibatch-size 128 --num-epochs 10 --method GE-binomial --slack -1 --autoencoder 0 --l2 0.0 --minibatch-balance 0.0625 --epoch-size 5000 --model resnet8 --units 32 --dropout 0.0 --bn on --unit-scaling 2 --ngf 32 --num-workers 24 --cross-validation-seed 1979721300 --radius 2 --num-particles 100 --device 0 --save-prefix=/home/user/processing/cryosparc_projects/empiar/S_protein/P22/J14/models/model -o /home/user/processing/cryosparc_projects/empiar/S_protein/P22/J14/train_test_curve.txt
[CPU: 216.1 MB] # Loading model: resnet8
[CPU: 216.1 MB] # Model parameters: units=32, dropout=0.0, bn=on
[CPU: 216.1 MB] # Loading pretrained model: resnet8_u32
[CPU: 216.1 MB] # Receptive field: 71
[CPU: 216.1 MB] # Using device=0 with cuda=True
[CPU: 216.1 MB] Traceback (most recent call last):
[CPU: 216.1 MB] File "/home/user/software/anaconda3/envs/topaz/bin/topaz", line 11, in <module>
[CPU: 216.1 MB] load_entry_point('topaz-em==0.2.4', 'console_scripts', 'topaz')()
[CPU: 216.1 MB] File "/home/user/software/anaconda3/envs/topaz/lib/python3.6/site-packages/topaz/main.py", line 148, in main
[CPU: 216.1 MB] args.func(args)
[CPU: 216.1 MB] File "/home/user/software/anaconda3/envs/topaz/lib/python3.6/site-packages/topaz/commands/train.py", line 641, in main
[CPU: 216.1 MB] image_ext=args.image_ext
[CPU: 216.1 MB] File "/home/user/software/anaconda3/envs/topaz/lib/python3.6/site-packages/topaz/commands/train.py", line 221, in load_data
[CPU: 216.1 MB] train_images = pd.read_csv(train_images, sep='\t') # training image file list
[CPU: 216.1 MB] File "/home/user/software/anaconda3/envs/topaz/lib/python3.6/site-packages/pandas/io/parsers.py", line 676, in parser_f
[CPU: 216.1 MB] return _read(filepath_or_buffer, kwds)
[CPU: 216.1 MB] File "/home/user/software/anaconda3/envs/topaz/lib/python3.6/site-packages/pandas/io/parsers.py", line 448, in _read
[CPU: 216.1 MB] parser = TextFileReader(fp_or_buf, **kwds)
[CPU: 216.1 MB] File "/home/user/software/anaconda3/envs/topaz/lib/python3.6/site-packages/pandas/io/parsers.py", line 880, in __init__
[CPU: 216.1 MB] self._make_engine(self.engine)
[CPU: 216.1 MB] File "/home/user/software/anaconda3/envs/topaz/lib/python3.6/site-packages/pandas/io/parsers.py", line 1114, in _make_engine
[CPU: 216.1 MB] self._engine = CParserWrapper(self.f, **self.options)
[CPU: 216.1 MB] File "/home/user/software/anaconda3/envs/topaz/lib/python3.6/site-packages/pandas/io/parsers.py", line 1891, in __init__
[CPU: 216.1 MB] self._reader = parsers.TextReader(src, **kwds)
[CPU: 216.1 MB] File "pandas/_libs/parsers.pyx", line 374, in pandas._libs.parsers.TextReader.__cinit__
[CPU: 216.1 MB] File "pandas/_libs/parsers.pyx", line 674, in pandas._libs.parsers.TextReader._setup_parser_source
[CPU: 216.1 MB] FileNotFoundError: [Errno 2] File /home/user/processing/cryosparc_projects/empiar/S_protein/P22/J14/image_list_train.txt does not exist: '/home/user/processing/cryosparc_projects/empiar/S_protein/P22/J14/image_list_train.txt'
[CPU: 216.1 MB]
Training command complete.
[CPU: 216.1 MB] Training done in 6.045s.
[CPU: 216.1 MB] --------------------------------------------------------------
[CPU: 216.1 MB] Traceback (most recent call last):
File "cryosparc2_worker/cryosparc2_compute/run.py", line 85, in cryosparc2_compute.run.main
File "cryosparc2_compute/jobs/topaz/run_topaz.py", line 359, in run_topaz_wrapper_train
assert len(glob.glob(os.path.join(model_dir, '*'))) > 0, "Training failed, no models were created."
AssertionError: Training failed, no models were created.```