Dear cryoSPARC users,
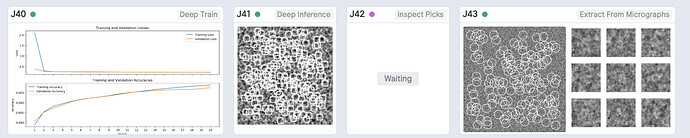
We have developed a new suite of deep-Learning based particle picking jobs in cryoSPARC that enable a user to train deep neural networks and then use those models to pick particles from micrographs. The first versions of these are available in a new beta version of cryoSPARC that can be optionally downloaded and used. We are looking for beta testers to try the new features and updated dependencies - anyone who wishes can upgrade to the beta version. We would appreciate any feedback users can provide, as well as bug reports or issues arising from the new dependencies.
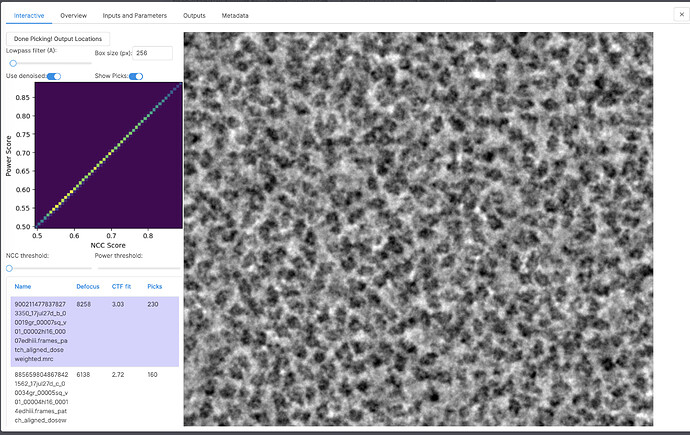
Specifically, the new version includes new job types: Deep Picker Train (BETA) and Deep Picker Inference (BETA). Deep Picker Train (BETA) allows the user to use a subset of micrographs and corresponding particles to train a deep neural network which can then be used in the Deep Picker Inference (BETA) job to pick particles from the remaining micrographs. You can find documentation on the new job types here: https://guide.cryosparc.com/processing-data/all-job-types-in-cryosparc/deep-picking/deep-network-particle-picker
These new jobs require updates to cryoSPARC’s core dependencies. These new dependencies will be automatically installed when you update to this version of cryoSPARC. The new installer bundles are substantially larger than before, so update may take some time.
If you do wish to update, you can do so from any version of cryoSPARC by running:
cryosparcm update --version=v2.16.0-deeppick_privatebeta
# or, for cryoSPARC live:
cryosparcm update --version=v2.16.1-live_deeppick_privatebeta
If you run into any issues on the deep picker and/or dependencies, please reply to this thread.
Thank you!