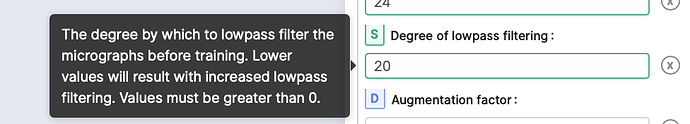
It’s not clear from the description what “degree of lowpass filtering” is. I would naturally assume it is a low pass filter of x Å, but the docs seem to contradict that, as they say a lower number gives more lowpass filtering. Is this right?
Hi @olibclarke,
We’re working on the incompatibility between inspect picks and deep inference. Also the lowpass filtering parameter has to be changed on our end. Currently the parameter is in wavenumber but we should change it to be a parameter in units of Angstroms.
Regards,
Jay Yoo
Great, thanks @jyoo! Also, there seems to be an issue with log output for Deep Train - sometimes it doesn’t output anything to the log after data augmentation is complete, until training is finished (when all the logs for training are output at once). This doesn’t really matter, but can lead the user to think that nothing is going on, especially given training can take several hours.
Cheers
Oli
Also the “shape of split micrographs” parameter - is this in downsampled pixels, or on the original scale of the micrograph?
Hi @olibclarke,
When the streamlog issue you mentioned occurs, try refreshing the browser and see if that restores the proper logging.
Regarding the “shape of split micrographs” parameter, this is in downsampled pixels.
Regards,
Jay Yoo
Thanks @jyoo, refreshing the browser does not help.
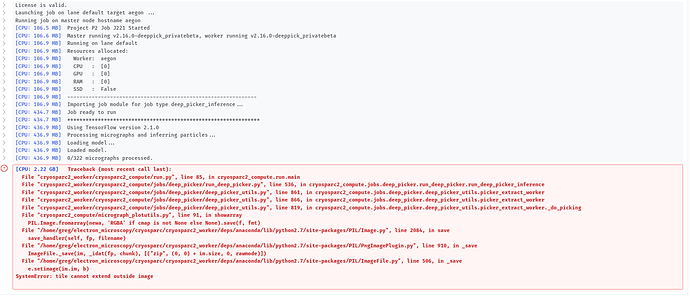
I’m getting a strange error when I try to run Deep picker inference on a model I’ve generated.
Single workstation, 2x2080ti GPU, 96 gig RAM, Ubuntu linux.
Output from cryosparc attached:
Hi @hansenbry, was the problem solved? I got the same error in cryoSPARC 3.0.1.
Hello @jyoo.
I am excited to try the Cryosparc deep picker but I am apparently having the same issue as @hansenbry and YYang:
Traceback (most recent call last):
File "cryosparc_worker/cryosparc_compute/run.py", line 84, in cryosparc_compute.run.main
File "cryosparc_worker/cryosparc_compute/jobs/deep_picker/run_deep_picker.py", line 255, in cryosparc_compute.jobs.deep_picker.run_deep_picker.run_deep_picker_train
AssertionError: No particles corresponding to input micrographs were found. Ensure that non-zero particle picks were input and that the particle picks are from the input micrographs.
I imported motion corrected micrographs but then used Cryosparc patch ctf. I tried connecting all possible micrographs objects to the Deep Picker job, and got the same error. Preprocessing seems to go OK so I think there is a real problem.
Full log
[CPU: 81.0 MB] Project P3 Job J200 Started
[CPU: 81.1 MB] Master running v3.0.1, worker running v3.0.1
[CPU: 81.3 MB] Running on lane lmb-gpu-nodes
[CPU: 81.3 MB] Resources allocated:
[CPU: 81.3 MB] Worker: lmb-gpu-nodes
[CPU: 81.3 MB] CPU : [0, 1, 2, 3]
[CPU: 81.3 MB] GPU : [0, 1, 2, 3]
[CPU: 81.3 MB] RAM : [0, 1, 2, 3, 4]
[CPU: 81.3 MB] SSD : False
[CPU: 81.3 MB] --------------------------------------------------------------
[CPU: 81.3 MB] Importing job module for job type deep_picker_train...
[CPU: 353.9 MB] Job ready to run
[CPU: 354.0 MB] ***************************************************************
[CPU: 442.3 MB] Using TensorFlow version 2.2.0
[CPU: 462.3 MB] Random seed used is 1956670233
[CPU: 462.3 MB] Processing micrographs and labels...
[CPU: 521.8 MB] Preprocessing over 4 processes...
[CPU: 521.8 MB]
[CPU: 521.8 MB] #################
[CPU: 521.8 MB] Process 1 Logs:
[CPU: 521.8 MB] 50/50 micrographs and corresponding labels processed.
[CPU: 521.8 MB] 50/50 micrographs split.
[CPU: 521.8 MB] Processing micrographs and labels done in 106.551 seconds.
[CPU: 521.8 MB] Percentage of positive labels is: 2.299%
[CPU: 521.8 MB]
[CPU: 521.8 MB] Splitting micrographs...
[CPU: 521.8 MB] Splitting micrographs done in 0.040 seconds.
[CPU: 521.8 MB] Augmenting data...
[CPU: 521.8 MB] 1250/1250 micrographs augmented.
[CPU: 521.8 MB] Augmenting data done in 51.106 seconds.
[CPU: 521.8 MB] #################
[CPU: 521.8 MB] Process 2 Logs:
[CPU: 521.8 MB] 50/50 micrographs and corresponding labels processed.
[CPU: 521.8 MB] 50/50 micrographs split.
[CPU: 521.8 MB] Processing micrographs and labels done in 109.702 seconds.
[CPU: 521.8 MB] Percentage of positive labels is: 2.478%
[CPU: 521.8 MB]
[CPU: 521.8 MB] Splitting micrographs...
[CPU: 521.8 MB] Splitting micrographs done in 0.039 seconds.
[CPU: 521.8 MB] Augmenting data...
[CPU: 521.8 MB] 1250/1250 micrographs augmented.
[CPU: 521.8 MB] Augmenting data done in 54.895 seconds.
[CPU: 521.8 MB] #################
[CPU: 521.8 MB] Process 3 Logs:
[CPU: 521.8 MB] 50/50 micrographs and corresponding labels processed.
[CPU: 521.8 MB] 50/50 micrographs split.
[CPU: 521.8 MB] Processing micrographs and labels done in 111.006 seconds.
[CPU: 521.8 MB] Percentage of positive labels is: 2.704%
[CPU: 521.8 MB]
[CPU: 521.8 MB] Splitting micrographs...
[CPU: 521.8 MB] Splitting micrographs done in 0.031 seconds.
[CPU: 521.8 MB] Augmenting data...
[CPU: 521.8 MB] 1250/1250 micrographs augmented.
[CPU: 521.8 MB] Augmenting data done in 73.694 seconds.
[CPU: 521.8 MB] #################
[CPU: 521.8 MB] Process 4 Logs:
[CPU: 521.8 MB] 50/50 micrographs and corresponding labels processed.
[CPU: 521.8 MB] 50/50 micrographs split.
[CPU: 521.8 MB] Processing micrographs and labels done in 111.486 seconds.
[CPU: 521.8 MB] Percentage of positive labels is: 2.057%
[CPU: 521.8 MB]
[CPU: 521.8 MB] Splitting micrographs...
[CPU: 521.8 MB] Splitting micrographs done in 0.029 seconds.
[CPU: 521.8 MB] Augmenting data...
[CPU: 521.8 MB] 1250/1250 micrographs augmented.
[CPU: 521.8 MB] Augmenting data done in 53.637 seconds.
[CPU: 521.8 MB] #################
[CPU: 521.8 MB]
[CPU: 521.1 MB] Traceback (most recent call last):
File "cryosparc_worker/cryosparc_compute/run.py", line 84, in cryosparc_compute.run.main
File "cryosparc_worker/cryosparc_compute/jobs/deep_picker/run_deep_picker.py", line 255, in cryosparc_compute.jobs.deep_picker.run_deep_picker.run_deep_picker_train
AssertionError: No particles corresponding to input micrographs were found. Ensure that non-zero particle picks were input and that the particle picks are from the input micrographs.
Hi @jwag,
Looks like you were running the deep picker training job. Where did you get the particles from for this training? Manual picks? blob picks? Are you sure the particles are picked on the same micrographs as you are passing to training?
Hi Ali. Thanks for getting back to me. Particles are from 2D Classification, originally picked with Filament Tracer. I tried using Micrographs output from Particle Extraction, Patch CTF and Import, all with same result. As per image attached, the Micrographs are the ones picked from.
An update - Topaz train begins training with the same inputs (still running). Also, job fails in the same way regardless of input into Number of micrographs to process.
Hello @jwag,
Any success with the Topaz Train job? Were the particles used for training the same as in your previous post (i.e. originally picked with the filament tracer)?
Best,
Michael
Hi @mmclean - yes the Topaz job succeeds with the inputs that fail for Cryosparc implementation.
Thanks
James
I am getting the same error! No particles corresponding to input micrographs were found. Ensure that non-zero particle picks were input and that the particle picks are from the input micrographs.
@jwag You succeeded in running deep picker?
No I didn’t - but I haven’t tried again.
Hi @tushancse, @jwag,
In cryoSPARC v3.2.0, we released a fix to the Deep Picker Train job that addresses this issue (it had to do with the job incorrectly matching particles and micrographs by their filenames instead of their unique identifiers, which might not be unique). Can you try updating and give it a go? cryosparcm update
Hi @stephan, we see the same issue of inspect picks output not working with Deep Picker in 3.2. Manual Picker works, but Inspect Picks shows zero particles