Dear all,
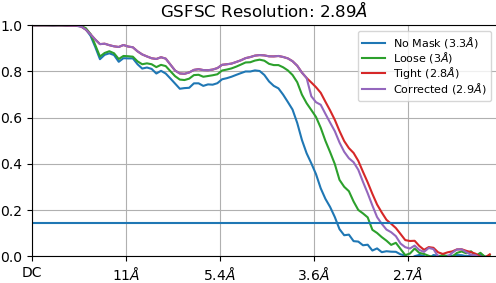
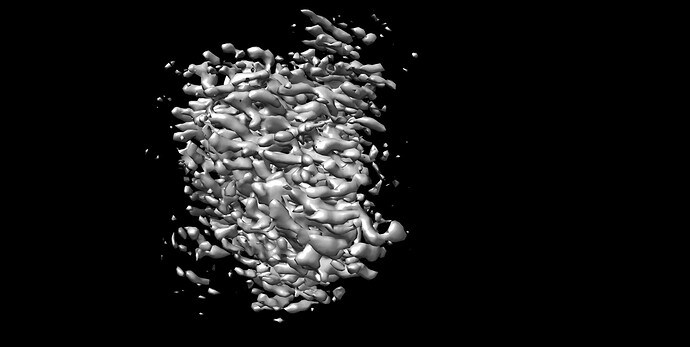
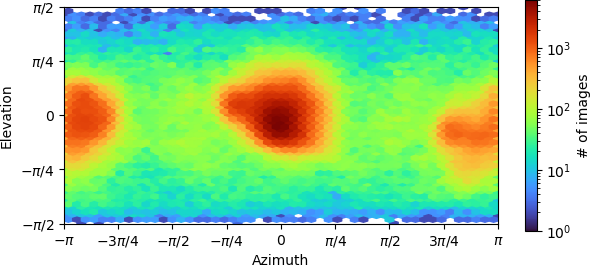
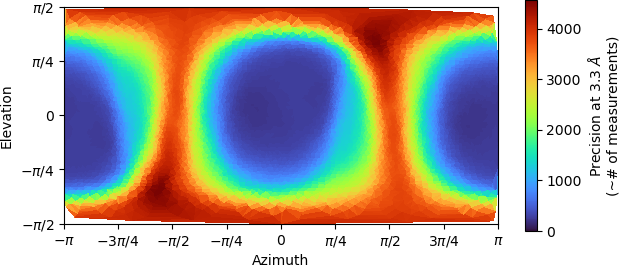
Recently we processed one CryoEM structure to 2.9 Armstrong from Cryosparc, the FSC curve looks ok. However, when we took a look in Chimera and Coot, the structure doesn’t really looks like that, we couldn’t identify a clear secondary structure; furthermore, if we were trying to fit the an alphafold prediction pdb into the map, we couldn’t fit in.
I wonder do you know how could this happen? Is there any ways we can do to improve the map?
Thanks!