Hi there,
I’m having some trouble trying to convert Euler angle for membrane-embedded proteins into cryoSPARC’s axis-angle orientation.
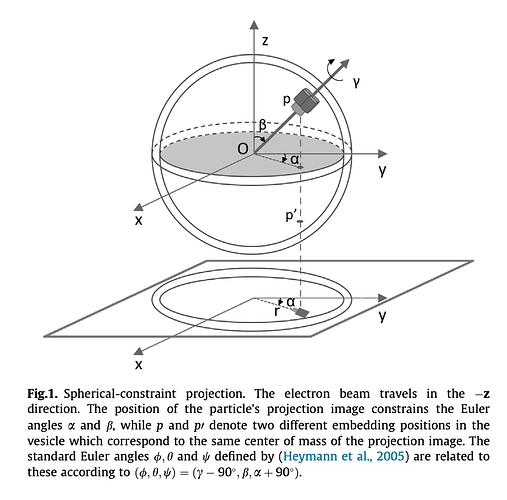
All my proteins are embedded into vesicles, and the subset I’m working with are such that they are only side-views (only on the sides of the vesicles, none at the top). I’m using the convention from Sigworth et al.'s 2014 paper, pictured below, to get a \beta = \pi/2 and estimating an \alpha. Then, I use the suggested conversion to Euler angles.
I should also add that I’m working with \phi \in [-\pi, \pi], \theta \in [0, \pi], \psi \in [ -\pi, \pi].
I then use Asarnow’s pyem/geom/convert.py (going from Euler → Rot Mat → Quaternion → Axis-Angle) to get axis-angle representations, and write these particles to a new cryosparc file. However, when trying to do backprojection with cryoDRGN (haven’t gotten to trying it out with cryoSPARC yet), I get garbage structures, with a centre sphere and noise around it. I tried with and without the --invert-data flag.
I do know that these angles are somehow correct, since I have tried reading from the cryosparc file, using Asarnow’s functions again, adding \pi/2 to the third Euler angle, rotating the images, and finally averaging them, getting a coherent structure.
Any help regarding my workflow would be greatly appreciated. Additionally, I have read both “How is “alignments3D/pose” encoded?” and “It’s time I admit I don’t know what Rodrigues vectors are” (unfortunately, cannot link these), but I’m still facing this issue.