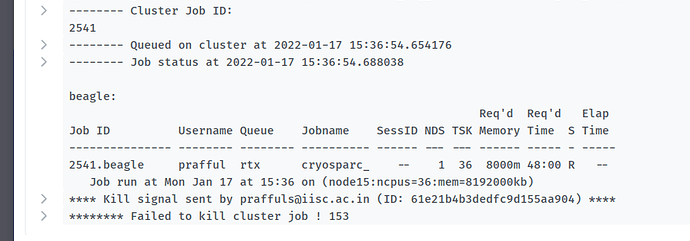
I tried installing cryosparc on pbs type of cluster and it installed successfully however when I launch any job the job is launched and it also shows that it is queued but when I check it on cluster using qstat command it does not show up in the queued list. When I try to kill the job it shows an error “Command ‘[‘qdel’, ‘2540’]’ returned non-zero exit status 153”.
I have attached the screenshot of the error
Welcome to the forum @prafful. Did you notice the mismatch between the job id in the text of your message (2540) and in the screenshot (2541)? In case that’s just a typo, can you find out more about the fate of job 2541?
Yes that was a typo. Job 2541 shows the same error “Command ‘[qdel’, 2541’]’ returned non-zero exit status 153”. The error log file shows the below error
Fatal Python error: _PyMainInterpreterConfig_Read: memory allocation failed
ValueError: character U+656e512d is not in range [U+0000; U+10ffff]
Current thread 0x00007f4483f9a740 (most recent call first):
/var/spool/pbs/mom_priv/jobs/2541.beagle.SC: line 40: seq: command not found
@prafful Does your cluster_script.sh template include a call to the seq command? In that case, is seq installed on the compute nodes and the output of dirname $(which seq) (on the compute nodes) in your $PATH?
Below is the script.sh file that I have used. Do you find anything wrong in it? It should be installed because other people are able to use cryosparc using the same compute nodes.
#!/bin/bash
cryoSPARC cluster submission script template for PBS
Available variables:
{{ run_cmd }} - the complete command string to run the job
{{ num_cpu }} - the number of CPUs needed
{{ num_gpu }} - the number of GPUs needed.
Note: the code will use this many GPUs starting from dev id 0
the cluster scheduler or this script have the responsibility
of setting CUDA_VISIBLE_DEVICES so that the job code ends up
using the correct cluster-allocated GPUs.
{{ ram_gb }} - the amount of RAM needed in GB
{{ job_dir_abs }} - absolute path to the job directory
{{ project_dir_abs }} - absolute path to the project dir
{{ job_log_path_abs }} - absolute path to the log file for the job
{{ worker_bin_path }} - absolute path to the cryosparc worker command
{{ run_args }} - arguments to be passed to cryosparcw run
{{ project_uid }} - uid of the project
{{ job_uid }} - uid of the job
{{ job_creator }} - name of the user that created the job (may contain spaces)
{{ cryosparc_username }} - cryosparc username of the user that created the job (usually an email)
What follows is a simple PBS script:
#PBS -N cryosparc_{{ project_uid }}_{{ job_uid }}
#PBS -l select=1:ncpus=9:ngpus={{ num_gpu }}:mem={{ (ram_gb*1000)|int }}mb
#PBS -o {{ job_dir_abs }}
#PBS -e {{ job_dir_abs }}
#PBS -q rtx
available_devs=“”
for devidx in (seq 0 3);
do
if [[ -z (nvidia-smi -i $devidx --query-compute-apps=pid --format=csv,noheader) ]] ; then
if [[ -z “$available_devs” ]] ; then
available_devs=$devidx
else
available_devs=$available_devs,$devidx
fi
fi
done
export CUDA_VISIBLE_DEVICES=$available_devs
{{ run_cmd }}
@prafful
Some points that may be helpful in further troubleshooting:
- Do you and the other users whose cryoSPARC jobs are correctly running share a single cryoSPARC instance, database, etc.?
- Does the template you posted match the active cryoSPARC configuration? You can use the
get_scheduler_target_cluster_script()function viacryosparcm clito find out. - Inspect job-related in- and output files, like
/var/spool/pbs/mom_priv/jobs/2541.beagle.SCand ensure all relevant commands are available on the worker node.
/var/spool/pbs/mom_priv/jobs/2541.beagle.SC: line 40: seq: command not foundsuggests thatseqis either not installed or not “in the path” on the scheduler-assigned worker node. Keep in mind that the path “seen” by a cluster job may differ from the “$PATH” that’s active upon direct login to the same worker node. - Reach out to your organization’s cluster support staff for help and suggestions.
The issue is solved now. I had installed Phenix earlier and cryosparc was using the Python from Phenix that was incompatible for cryosparc. This may have occured because Phenix comes up first chronologically and in Phenix it searches for python and detect that version.
I could figure this out in the error and job log files.
Thanks for the help!