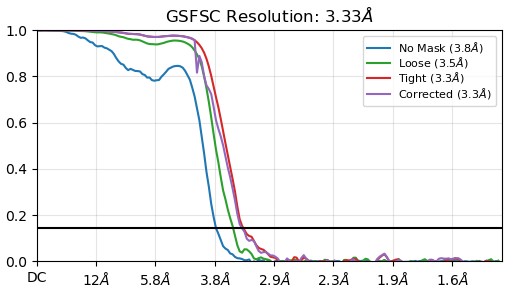
Hey there!
I was hoping that someone with more experience moving between Cryosparc and Relion might have some input for me.
I’m currently working with a relatively complex, a protein plus inhibitor of ca. 80 kDa together, no symmetry.
After looking at the sample with a screening microscope, I managed to get a decent map at maybe 8-10 Å resolution, then collected data on a Krios and, after failing to get good classes with blob pickers, used a template I created from NU refinement with the screening microscope data.
2D classifications improved a lot, although there was an orientation bias. I did some initial refinements, then used a resulting good map with four decoy volumes in heterogeneous refinement. Went from 6.3 mil particles to 1.9 mil after NU refinement with an okay-looking model with obvious overfitting issues and preferred orientation issues (cFAR of 0.27). Followed up with 3D classification and found a good class with 220k particles leading to a map with a final resolution of 3.33 Å after NU refinement and cFAR of 0.60 (including particle polishing and global ctf refinement), so less orientation biased if I am understanding that correctly.
Since nothing I tried since then improved things, I wanted to move to Relion to see if post-processing and blush regularization there might yield any more improvements, but that’s where I am stuck.
I’ve imported and processed the micrographs in Relion, extracted the particles used for NU-refinement after the 3D-classification (confirmed that they’re being extracted correctly in Relion), and then tried both initial modelling as well as 3D auto-refinement, with pretty terrible results.
Initial modelling results in maps of 8 to 10 Å, according to Relion, that just look like non-converging junk or noise. 3D refinement sometimes does the same, or leads to volumes looking vaguely like my cryosparc results but with additional noise mixed in, and are stuck around 14 to 16 Å resolution (and look like it).
Using relion_reconstruct results in a volume with the right orientation and very roughly a similar shape, but also if low-pass filtered to maybe 20 Å.
Reducing off-set steps and range or angular sampling doesn’t help.
I’ve had others confirm that the cryosparc map looks decent, and ModelAngelo can build maybe 70% of the model with it as input, so I am reasonably sure those results are trustworthy.
Does anyone have any idea what I could do to improve the processing in Relion? Or is this just one of the random cases where thing’s don’t work somehow?
I’d be grateful for any input!
Best,
Leon