I’ve long been puzzled by the pros and cons of providing both solvent and focus masks for 3D classification, compared with using only one mask covering the ROI as is implemented in Relion. I can understand the former is a cheap alternative to particle subtraction, in the sense that it accounts for the signal and CTF of the region outside the ROI that bleed into it. In my experience, providing both masks barely separates the particles. The particle distribution is virtually uniform and all the class averages look highly similar. In contrast, if I provide just the solvent mask covering the ROI, the particles are much more efficiently separated.
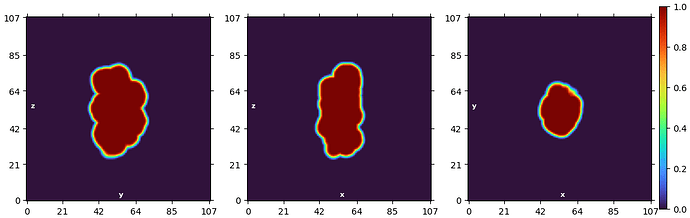
To put it in more context: I have 4 million particles. Each particle is a segment on a filament. The solvent mask is a soft mask covering the 5 subunits in the center (43 kD for each subunit, 215 kD in total). The focus mask covers the ROI, where we anticipate to see the binding of a 7 kD domain. The solvent mask is generous so that it completely covers the focus mask. About 5% particles are bound by the 7 kD domain and they are the ones I’m hoping to find by 3D classification. Below is how the masks look like:
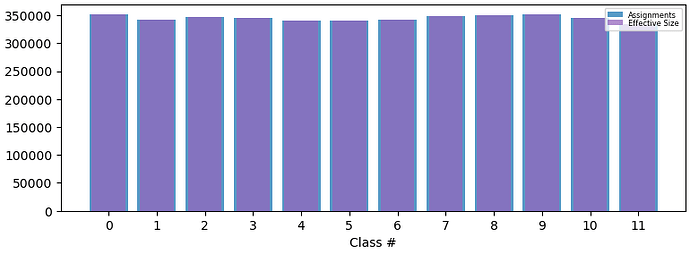
Below is the final particle distribution with both masks provided:
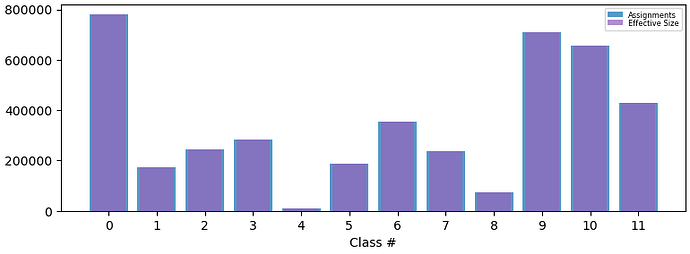
However, if providing just the solvent mask covering the ROI, that is using the focus mask from the previous job as the solvent mask, the particles are much better separated:
Right now running 3D classification with just solvent mask seems working fine. But I’d like to understand why using both masks performs worse, as it’s counterintuitive.
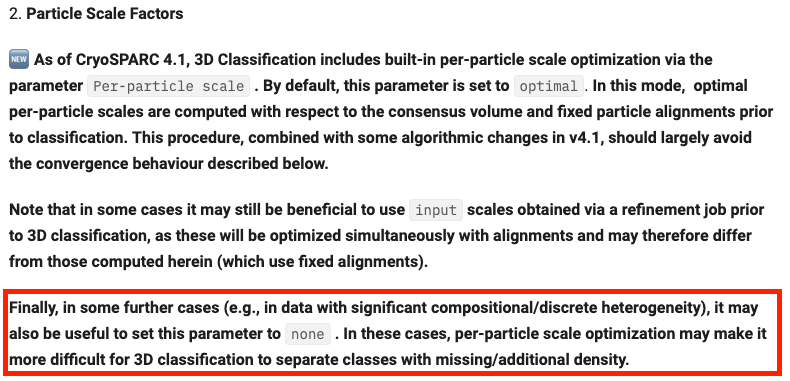
On a related note, how is the per-particle scale computed if just a small solvent mask covering the ROI is used? I’ve noticed setting the parameter to none consistently performs better than optimal. The documentation says setting it to “none” is only recommended for very small particles or strange cases. I was wondering what are those “strange cases”? Could it be more specific? And in the case of using only the solvent mask, does “very small particle” actually refer to very small mask?