I have started to have problems in v3.2.0 with some homogeneous refinement jobs failing with an AssertionError: No match for particles.blob as the job completes. It can be solved by rerunning the job but is a little irritating.
This is also an error message which is present when I try to run an automatic intermediate file cleanup, similar to previous reports: https://discuss.cryosparc.com/t/clearing-intermediate-results-hangs/4954/16
This doesn’t seem to be a permission problem as shared in that thread.
Hi @donaldb, can you please paste the full traceback from the homogeneous refinement job that failed with this error?
Hi @spunjani.
This is actually from a non-uniform refinement but they are the same error for both types, although I had already deleted the job:
Traceback (most recent call last):
File "cryosparc_worker/cryosparc_compute/run.py", line 84, in cryosparc_compute.run.main
File "cryosparc_worker/cryosparc_compute/jobs/refine/newrun.py", line 68, in cryosparc_compute.jobs.refine.newrun.run_homo_refine
File "/raid/benond/Cryosparc_v2_Install/cryosparc2_worker/cryosparc_compute/jobs/runcommon.py", line 550, in load_input_group
dsets = [load_input_connection_slots(input_group_name, keep_slot_names, idx, allow_passthrough=allow_passthrough) for idx in range(num_connections)]
File "/raid/benond/Cryosparc_v2_Install/cryosparc2_worker/cryosparc_compute/jobs/runcommon.py", line 550, in <listcomp>
dsets = [load_input_connection_slots(input_group_name, keep_slot_names, idx, allow_passthrough=allow_passthrough) for idx in range(num_connections)]
File "/raid/benond/Cryosparc_v2_Install/cryosparc2_worker/cryosparc_compute/jobs/runcommon.py", line 521, in load_input_connection_slots
dsets = [load_input_connection_single_slot(input_group_name, slot_name, connection_idx, allow_passthrough=allow_passthrough) for slot_name in slot_names]
File "/raid/benond/Cryosparc_v2_Install/cryosparc2_worker/cryosparc_compute/jobs/runcommon.py", line 521, in <listcomp>
dsets = [load_input_connection_single_slot(input_group_name, slot_name, connection_idx, allow_passthrough=allow_passthrough) for slot_name in slot_names]
File "/raid/benond/Cryosparc_v2_Install/cryosparc2_worker/cryosparc_compute/jobs/runcommon.py", line 511, in load_input_connection_single_slot
output_result = com.query(otherjob['output_results'], lambda r : r['group_name'] == slotconnection['group_name'] and r['name'] == slotconnection['result_name'], error='No match for %s.%s in job %s' % (slotconnection['group_name'], slotconnection['result_name'], job['uid']))
File "/raid/benond/Cryosparc_v2_Install/cryosparc2_worker/cryosparc_compute/jobs/common.py", line 615, in query
assert res != default, error
AssertionError: No match for particles.blob in job J116
Thanks a lot.
Hey @donaldb,
What are the inputs to your homogeneous refinement/non-uniform refinement jobs? Are they particles from an Import Particles job?
The input is an imported relion stack.
The strange thing is if I clear and restart the job I get the same error or if I clone the job but if I make a fresh job and use the same input it can work fine with the same import job.
Hi @donaldb,
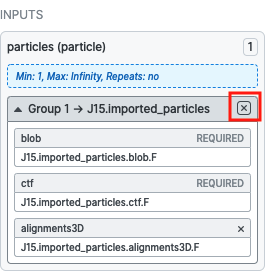
When you first create an “Import Particles” job, the output groups don’t contain specific output result slots like particles.blob or particles.ctf. Until the job is actually finished, the job doesn’t know whether the .star file you’ve specified to import has references to the blob, ctf, location, alignments2D or alignments3D data for the particle stack; it has to find out by reading the star file itself, which it can’t do when you “build” the job.
I suspect what happened here is that you connected the outputs of the “Import Particles” job to the “Non-Uniform Refinement” job before the “Import Particles” job completed, causing only the particles output result group to be connected, and not the particles.blob result slot, which is one of the required inputs.
This also makes sense, based on how the cryoSPARC job builder works: when you clone a job, it takes an exact copy of the job it came from, including it’s inputs, which in this case didn’t have particles.blob.
You can get around this by deleting the inputs to the “Non-Uniform Refinement” job and re-connecting the imported_particles output result group from the “Import Particles” job (which, assuming it’s completed, will now have all the result slots from the .star file).
This is essentially what you did when you created a new job and reconnected the inputs.
All to say, this is how it is  . It’s technically not a bug but definitely not the best way to do this, especially since it breaks the common workflow of being able to queue up a string of jobs before any parent jobs have completed. We’re currently working on a redesign of the cryoSPARC job builder, and this is something that will be addressed by it.
. It’s technically not a bug but definitely not the best way to do this, especially since it breaks the common workflow of being able to queue up a string of jobs before any parent jobs have completed. We’re currently working on a redesign of the cryoSPARC job builder, and this is something that will be addressed by it.
1 Like
Ah. Thanks for clarifying that. I think I was trying to start some jobs ASAP before the import had quite finished. It is quite an irritating problem, as it often appears when the job is writing out the final files after a few hours of computation.
I would definitely be in favour of a redesign of this, as it would be great if you could queue up the output from an import before it has fully finished and if it could ‘reserve judgement’ on the information in the import. The import jobs can often take quite some time on the Reading MRC file headers to check shape... step. Is it strictly necessary to check every mrc file from a relion extract job? This can be a lot of files for a large dataset, as the file convention from relion is one .mrcs file for each micrograph, which can easily end up with >10k files.
Thanks a lot.
Hey @donaldb,
That makes sense.
Not necessarily, we will add an option in this job to “Skip Header Check” so we don’t have to read the header of every file. This prevents catching any corrupted files early on, but will reduce the time it takes to import the particles.