Hello,
I am trying to extract particles from micrographs, but it always failed, even though I have tried to rescale the box size, bin 2 or bin 4. The total particle numbers were about 7 millions.
Any help is appreciated.
Hi @Jessica-Cai,
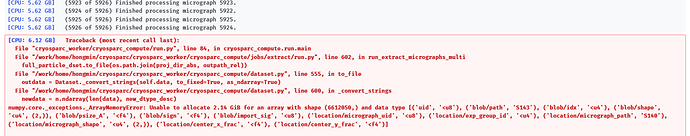
This “Unable to allocate” error happens when the Extract job tries to save the particle output meta-dataset after extraction is complete. This dataset does not contain the actual particle images but references to where those images are found on disk (which is why the memory error happens regardless of particle box size). It seems the job process is having trouble allocating the memory required to save it.
The best way to fix this is to reduce the amount of particles you’re providing as input for the Extraction job. You can do this with one of the following strategies:
- Stricter filters on the Inspect Picks job to reduce the amount of non-particles
- Split up the particles after picking (before extraction) and then extracting in separate jobs
For the second option: After Picking and Inspecting, provide the particles as input to a Particle Sets Tool job. Set the num. batches parameter to 4 or so. Set the batch size parameter to the number of particles divided by 4 (or whatever number you specific for the num. batches), rounded down to a whole number. Then run the job.
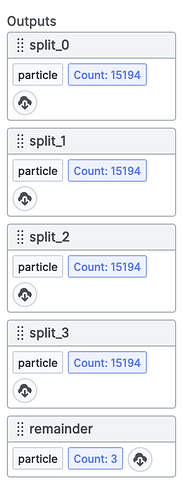
When complete you should have 4 split_ outputs that look like this:
Where each output represents a subset of the total particle dataset.
Create four (or however many splits there are) Extract Particles job for each split_ output as its particles input and all the micrographs. Run those jobs. Once complete, you can re-combine all the extracted particles in the next job by dragging all the extracted outputs into the same input slot for the next job.
Hope that helps, let me know if you run into any trouble with those steps.