Hi all,
I’m currently facing a difficulties in reconstructing membrane protein particles, which has little or no features outside the detergent micelles.
(apologies in case there are any inaccuracies in the use of terms or misunderstanding  )
)
The protein I am working on has an size of ~50kDa and the longest diameter is ~100Å.
Inspired from the other works and to see the different conformation, I am attempting to solve the protein structure without fiducial markers.
Fortunately, I could obtain ~20,000 micrographs which seems promising.
(on a Krios with energy filter with a raw pixel size of 0.664.)

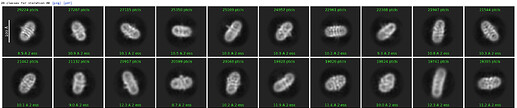
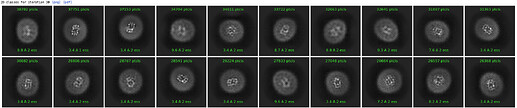
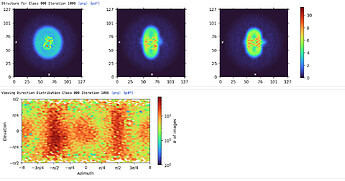
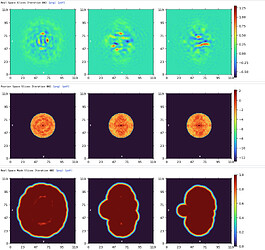
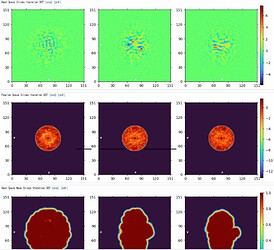
Here are parts of my 2D & Ab-initio (300k particles, 12~9Å, batch size is bigger than 1000) results.
Ab-initio map showed well defined TM domains with a small ICL outside of the micelle.
=========
Where I got stuck is the next step of this initial model…
Considering the TM densities and relatively weak micelle signals, I hope particles to be either classified or aligned correctly in refinement jobs. However none of the refinement jobs, including heterogenous-, homogenous-, NU- refinements resulted in successful outcomes. I’ve attempted homogenous/NU-refinement with lower initial resolution parameters (2) heterogenous refinement with force hard classification ‘on’ and increasing batch sizes (2) further ab-initio with multiple classes and increasing resolution ranges (3) 3D classification with the other junk volumes. with various parameters.
All I could see is greenish real space slices with little features of my protein.
==========
Is it because the signal is not sufficient for particle alignment in refinement jobs? or
could it be that the particles are not aligning well because they are not fully separated yet?
I am now producing the sample with smaller micelle to increase the TMD signals, however, still believe that there is poteintial to obtain better results from this dataset.
What can I do next?
Any advice is appreciated.
Thanks
U.Kim
2 Likes
A number of possibilities, but to start with your box looks very small for the particle - have you tried extracting in a larger box?
2 Likes
I am interested in how you solve this riddle as I have the same problem (I have virtually the same 2D classes as you), but I can’t even get a good ab-initio so I’m even behind.
Just a word on the detergent belt (not a micelle). If you go to a smaller size detergent, you might not get a smaller belt, this is because the protein needs a certain amount of amphipathic molecules to shield its hydrophobic layer, and this number is very different from the micelle aggregation number. If you were in DDM, you might have between 300-400 molecules around your protein, and if you switch to OG you will end-up with say 700 molecules of OG with a similar belt. See the gallery page of the detbelt server for examples of proteins where detergents have been quantified (https://detbelt.ibcp.fr/gallery).
The only case where I was able to reduce the belt size is the mixture of DDM and Cholate in 3:1 or 1:1 ratio, it also thermostabilized the protein.
You might give it a try, but no guaranty your protein will like it (it’s protein dependent).
2 Likes
It does look great! I have some experience in it and I agree with olibclarke, your box size is very small. I work on a protein that also 50 kDa, and I found that anything below 360 px limited my resolution to some degree and going above did not help further.
And yes, your approach sounds very reasonable, keep on doing heterogenous refinements and ab-initio with multiple classes and sort out particles until your resolution does not improve anymore.
1 Like
Hi @olibclarke Thanks for the reply!
I remember reading your post mentioning the Henderson and Rosenthal (2003) paper about it. 
Absolutely, I started with particles in a larger boxes (400px) and then tested even bigger ones (480px).
The second map is the one from the particles in 480px boxes.
Things weren’t that different as you can see.
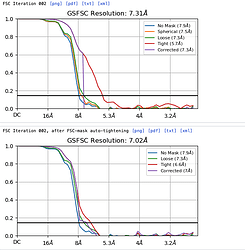
The initial resolution was 12 or less and also tried changing some mask parameters.
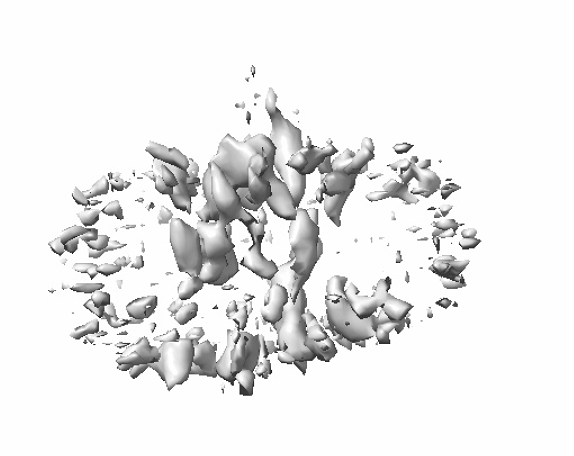
I gradually changed to smaller boxes during ab-initio jobs since the maps using larger boxes showed less discernible TMDs than the that of the smaller ones.
Thanks again,
U. Kim
Hi, @vincent
Thanks for clarifying the word. Mine is now surrounded by MNG-type detergents, and I was going to try DDM and ß-NG.
I didn’t know that cholate could influence the detbelt size, think I need to try.
Thanks for letting me know.
What is your final resolution set to? And secondly, how many particles do you have here? Did you try to run it with more classes like 4 or 6?
You can also try local refinement with a mask on the protein region excluding the micelle and a moderate lowpass filter. Getting thru the micelle in a case like this is difficult. I wonder if relion’s new blush refinement might produce a different result.
Also, larger box size, like Oli said. perhaps 1.5-2x larger
![]() )
)