Just revisiting this - I am still a bit puzzled. I have a single script that works, and I have it set up to make two jobs - one with a positive and one with a negative adjustment to defocus:
from pathlib import Path
import json
import re
import argparse
import numpy as np
from scipy.spatial.transform import Rotation as R
from cryosparc.tools import CryoSPARC
# Parse command-line arguments
parser = argparse.ArgumentParser(
description="Apply analytic defocus adjustments to CryoSPARC particle sets"
)
parser.add_argument(
"--project", "-p",
required=True,
help="CryoSPARC project UID (e.g., P10)"
)
parser.add_argument(
"--workspace", "-w",
required=True,
help="CryoSPARC workspace UID for new jobs (e.g., W3)"
)
parser.add_argument(
"--job", "-j",
required=True,
help="CryoSPARC job UID for the volume alignment job (e.g., J244)"
)
parser.add_argument(
"--instance-info", "-i",
default=str(Path.home() / 'instance_info.json'),
help="Path to CryoSPARC instance_info.json"
)
args = parser.parse_args()
# Load connection info
info_path = Path(args.instance_info)
with open(info_path, 'r') as f:
instance_info = json.load(f)
# Connect to CryoSPARC
cs = CryoSPARC(**instance_info)
if not cs.test_connection():
raise RuntimeError("Could not connect to CryoSPARC server")
# UIDs from arguments
PROJECT_UID = args.project
WORKSPACE_UID = args.workspace
VOL_ALIGN_UID = args.job
# Load project, job, volume, and particles
project = cs.find_project(PROJECT_UID)
job = project.find_job(VOL_ALIGN_UID)
volume = job.load_output("volume")
particles = job.load_output("particles")
# Map center (Å)
apix = volume['map/psize_A'][0]
map_size_px = volume['map/shape'][0]
map_center = (map_size_px * apix) / 2
# Determine new center shift (Å)
if 'recenter_shift' in job.doc.get('params_spec', {}):
s = job.doc['params_spec']['recenter_shift']['value']
m = re.match(r"([\d.]+),\s*([\d.]+),\s*([\d.]+)\s*(px|A)?", s)
if not m:
raise ValueError(f"Cannot parse recenter_shift: {s}")
center_vals = np.array([float(m.group(i)) for i in (1,2,3)])
new_center_A = center_vals * (apix if m.group(4) != 'A' else 1)
print("Center calculted from params.")
else:
log_entries = cs.cli.get_job_streamlog(PROJECT_UID, VOL_ALIGN_UID)
text = next(x['text'] for x in log_entries if 'New center' in x['text'])
m = re.search(r"([\d.]+),\s*([\d.]+),\s*([\d.]+)", text)
new_center_A = np.array([float(m.group(i)) for i in (1,2,3)]) * apix
print("Center extracted from log.")
delta = new_center_A - map_center
# Compute per-particle defocus shift
dz_vals = []
for row in particles.rows():
Rmat = R.from_rotvec(row['alignments3D/pose']).as_matrix()
beam_dir = Rmat[:, 2]
dz_vals.append(float(np.dot(beam_dir, delta)))
dz_array = np.array(dz_vals)
mean_dz = dz_array.mean()
std_dz = dz_array.std()
# Create adjustment jobs
for sign, label in [(1, 'plus'), (-1, 'minus')]:
parts = job.load_output('particles')
for row, dz in zip(parts.rows(), dz_array):
row['ctf/df1_A'] -= sign * dz
row['ctf/df2_A'] -= sign * dz
title = f"Defocus-adjusted_{label}"
out_job = project.create_external_job(
workspace_uid=WORKSPACE_UID,
title=title
)
out_job.add_input(type='particle', name='input_particles')
out_job.connect(
target_input='input_particles',
source_job_uid=VOL_ALIGN_UID,
source_output='particles'
)
out_job.add_output(
type='particle', name='particles', passthrough='input_particles',
slots=['ctf'], alloc=parts
)
out_job.save_output('particles', parts)
out_job.log(f"Applied analytic Δz {label}: mean={mean_dz * sign:.2f} Å, std={std_dz:.2f} Å")
out_job.stop()
print(f"Created job '{title}'")
print("All adjustment jobs created.")
I expected that flipping the sign would only be necessary if the handedness of the reconstruction was inverted - but it doesn’t seem to be the case.
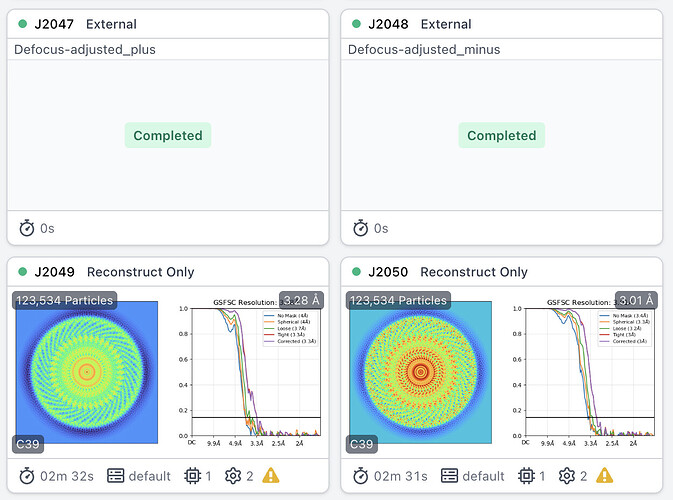
For example, for the vault particle, a negative adjustment is required for subparticles at the ends of the vault in order to improve resolution, while a positive adjustment is required for subparticles on the side of the vault (displaced from the Cn axis).
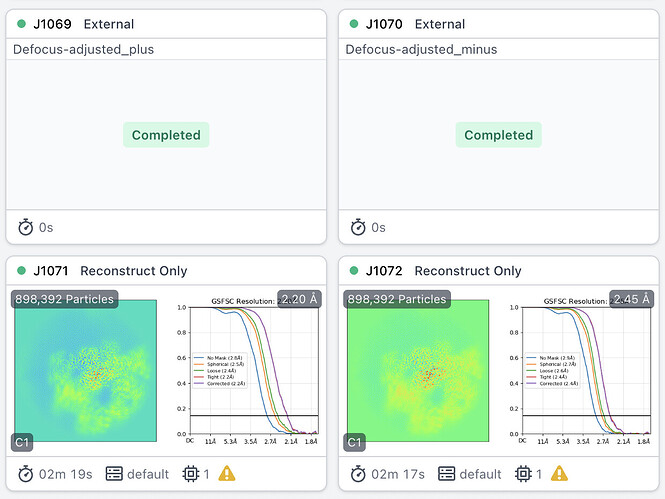
Similarly a positive adjustment is required for adjusting the defocus of subparticles at the corner of the ryanodine receptor. So I feel there must be a bug here somewhere, but I can’t see it for the life of me - the math should be the same no matter what particle/symmetry one is dealing with, and it certainly should be the same for different subparticles in the same assembly…
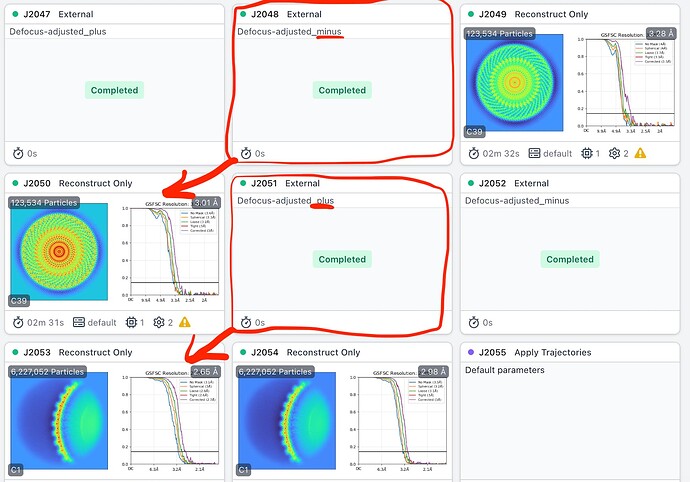
In practice maybe this doesn’t matter - I try both and see which one gives a resolution improvement after reconstruction - but I worry that perhaps it is applying the correct adjustments for some subparticles in a set, but not for others. Perhaps something to do with how the poses of symmetry expanded particles are specified?
Vault cap:
RyR corner:
Cheers
Oli