I tried that but it still failed with the same error, whether I had the input/output args at the start or the end…
EDIT: Okay, I think I found the issue @DanielAsarnow - it only works if I explicitly specify the pixel size with --apix - otherwise it fails with this error, even if I specify the box size.
Hmm - but now if I import this particle stack into cryosparc with the source mics connected, I get this error:
Traceback (most recent call last):
File "cryosparc_master/cryosparc_compute/run.py", line 115, in cryosparc_master.cryosparc_compute.run.main
File "/home/exx/cryosparc/cryosparc_worker/cryosparc_compute/jobs/imports/run.py", line 466, in run_import_particles
assert len(output_errors) == 0, "The following outputs were produced but not expected: " + ", ".join(output_errors)
AssertionError: The following outputs were produced but not expected: location
EDIT: Ok, got past that error by disabling strict output checking, and now it seems to be behaving, but the particle coords don’t seem to quite match the micrographs… the particles were downsampled, perhaps that is causing some issue with coordinate conversion in subparticles.py? @DanielAsarnow if the particle pixel size is different from the micrograph pixel size, will this cause a problem in the output coords?
EDIT: Ah got it! Needed --inverty in csparc2star.py. Working now! Only remaining issue is that I lose the per-group beam tilt etc upon import/export… I wish there was a more granular way to replace CTF information using the expanded inputs, e.g. a separate slot for defocus, tilt, trefoil etc… @rwaldo is there some way to copy over just the tilt/trefoil values using CS tools??
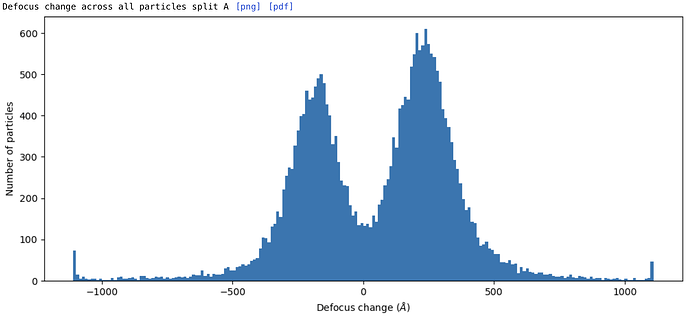
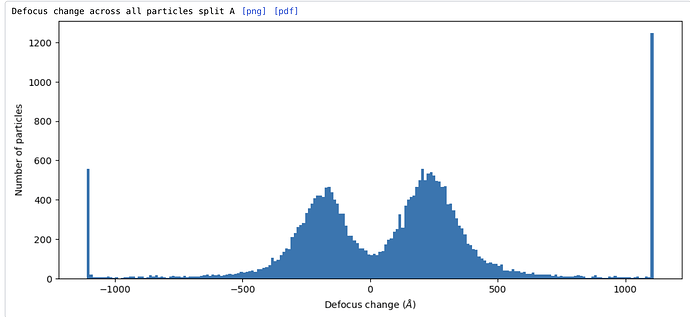
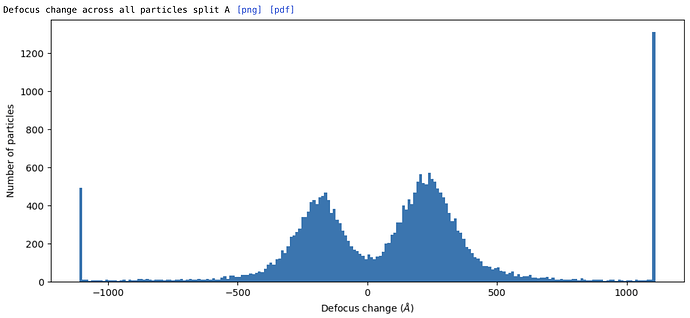
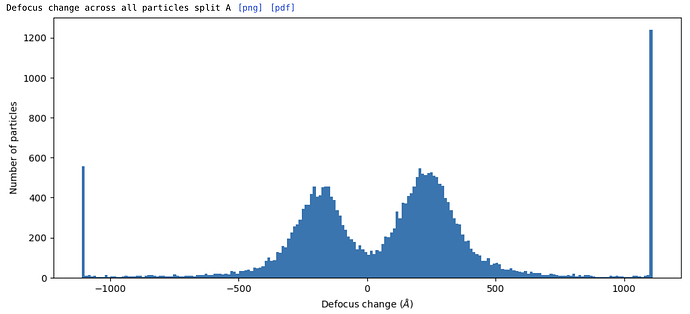
EDIT: Sort of working but not quite. The particle positions are still off when comparing to the original subparticle extraction in csparc - by perhaps 1/3 of the new subparticle box size, in a manner that seems like the z-value of the target is off somehow. Confirmed - the Z-value is offset by ~1/2 the subparticle box, but x and y are fine. Calculating the offset after extracting the mis-centered subparticles and adding it to the z-value of the target in subparticles.py (which gives a Z value larger than the box size in Å!) gives correctly centered subparticles. That does make me wonder whether the applied offsets to subparticle defocus will be correct, though…
EDIT: The resolution gets worse with --adjust-defocus in subparticles.py, which makes me think that the applied defocus changes are wrong - perhaps inverted? The hand of the initial reconstruction is correct, as is the reconstruction of the subparticles, so that isn’t the issue. Hmm. Tried using the --invert flag in subparticles.py, but the resolution is still worse than without --adjust-defocus (3.46 vs 3.29 Å). Resolution improves with per-particle CTF refinement, so I expect it ought to also improve when using --adjust-defocus instead, if the adjustments are correct.