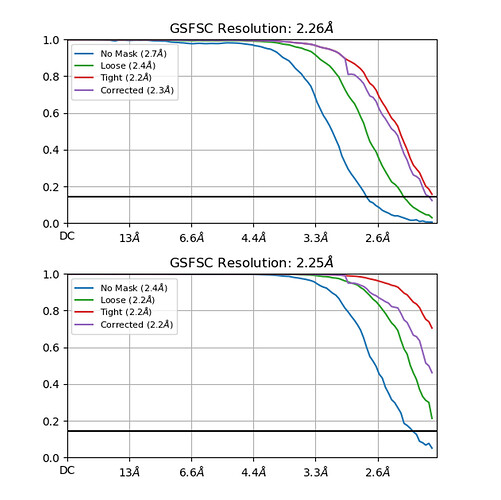
I am new to cryo-EM and cryosparc. When I processed my protein particles in C1 symmetry using Non-uniform refinement, I obtained the abnormal GSFSC curves shown below. In the areas shown, the curve seems to be particularly steep. At the same time, I also specify another Non-uniform refinement under D4 symmetry, which is even more serious. Does this curve mean that refine failed and the results are unreliable?
How does the map look? Can you see clear features consistent with the quoted resolution?
Without any additional information, it looks very good - your refinement is hitting the Nyquist limit imposed by your pixel size (or binning).
If your particles are binned, you might consider refining with unbinned data.
If not, you might consider looking at post acquisition super resolution (calling @rbs_sci & @alexjamesnoble!) - https://www.biorxiv.org/content/10.1101/2023.06.09.544325v1
As @olibclarke says, first check if the map looks “real” for the resolution. At 2.2Å you will want clear side chains and excellent secondary structure. If those a present, if using binned particles, unbin them. ![]() If particles are not binned, it does seem like PASR might be of use.
If particles are not binned, it does seem like PASR might be of use. ![]()
or
Thank you very much. I checked my volume and it should be fine. I plan to use PASR to try and deal with it later.
Nice. ![]() PASR is pretty easy and self-explanatory, but let me know if you have any questions.
PASR is pretty easy and self-explanatory, but let me know if you have any questions. ![]()