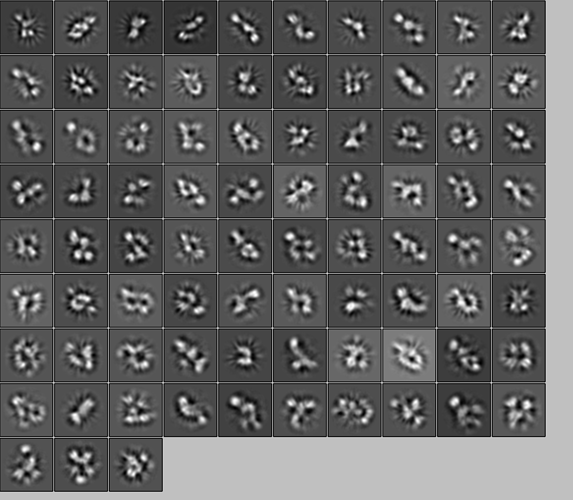
Hi,
I sent a sample of my complex to be analyzed by Cryo-EM, unfortunately, I don’t have a lot of experience with Cryo, but after they processed the data until the stage of ab initio, they told me that the sample was very heterogeneous.
The purification process showed really nice single peaks and SDS-PAGE showed purity around 95 % of purity.
Here is the snapshot of the models:
Could someone give me some insight if is possible to do the refinement having this data? I see that some structures in the picture look more or less the same, but with different conformations and orientations.
Thank you for your help,
Ismael
This does not look great and it would take a considerable effort from an expert to disentangle as you’ve described. They did not suggest your protein was impure, rather the same protein does not adopt 1 shape and has heterogeneous conformation. This you won’t see on SDS page since it’s a denaturing experiment. They’re likely saving you tons of effort from trying to solve structure(s) from this data. I would suggest a binding partner cofactor antibody oligomer-inducer, something to change it to 1 uniform constant shape. Or take the analysis at face value, you now know your protein is flexibly tethered together and that may inform on function.
1 Like