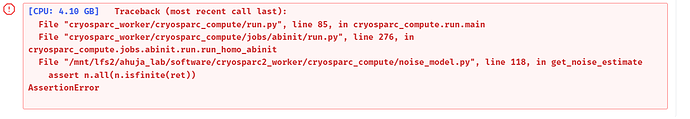
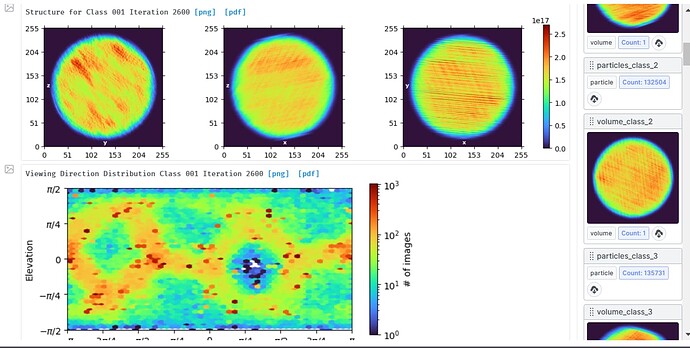
Hi, I am running v3.3.2 cryoSPARC, and after increasing my particle size, the ab initio jobs keep throwing the assertion error. The classes also gets very noisy and nonsensical sometimes, please see the images attached.
Welcome to the forum @tshi .
Please can you describe the steps involved in
?
Which inputs and/or parameters were changed compared to a previous successful ab initio reconstruction run, if there was one?
The previous successful runs had a extraction size of 600, fourier cropped to 256. The current run had the 1/2 cropping during motion correction, and a extraction size of 1200 and cropped to 300. There isn’t any change in the ab initio except for maybe number of classes.
Was that without cropping during motion correction?
That’s correct.
However, an ab initio job just now, from extraction size 2048 and fourier crop of 512 also just failed due to assertion error.
Did you confirm that the combination of F-croppings during motion and particle extraction results in a reasonable box extent (in Ångstroms), for example by finding interpretable 2D class averages with a reasonable particle-to-box size ratio?
I will run that right now to check.
However, other extractions without f-cropping yielded good 2D classes but still had the ab initio error.
Can I ask what the assertion error originate from in the software?
This particular AssertionError is supposed to catch “seriously misbehaving” calculations as indicated by the occurrence, with various causes, of non-finite or non-numerical values.
Would you be able to share images of those 2D class averages?
What prompted you to increase the box area?
As my previous box size did not capture the entirety of my complex, the box size was increased. As my complex is long, it required a fairly large box to have 2D classes capturing the entirety of it.
I previously thought that binning to 516 (not a power of 2) was causing it, but I re-ran extraction and binned to 512, but the same error persisted.
Are the complex’s projections expected to be much longer than they are wide, and perhaps more amenable to filament-type micrograph processing?
I will give that a shot.
Is there anything else I can do to troubleshoot the ab initio error? It still seems to me that ab initio remain the fastest and simplest way for me to obtain the final map.