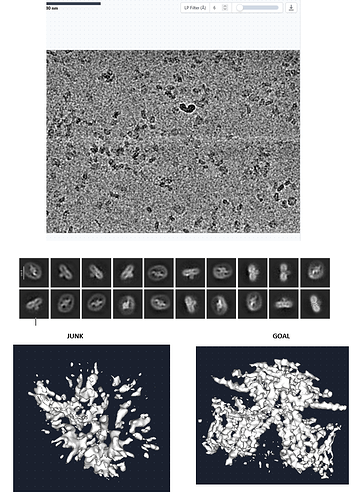
Dear all,
I am new in structural biology and Cryo-EM. I have been trying to acquire the structure of my protein for quite some time with no success (internal membrane protein in detergent micelle with small cytosolic domain). I have about 50.000 particles after strict inspection following template picking using older templates (original particle number was over 6 million from 20.000 micrographs). I managed to get good 2D classes after multiple rounds of 2-D classification using various settings. At the moment I am unable to reconstruct a 3D volume of my protein inside the detergent micelle. I have tried different Ab-initio settings as well as heterogeneous refinement again using an already solved structure of this protein, but nothing seems to work. The problem is that I get some extra density inside the micelle but it ends up being just junk. I can see parts of my protein in 2D but ab-initio can not align the different orientations to construct a complete 3D structure. Moreover, I have performed 3 session using the same microscope and conditions (TitanKrios 3Gi) and in all cases I was able to reach until the 2D classes stage, which makes me think there is something systematic that is wrong.
Could someone please suggest some good settings for Ab-initio or heterogeneous refinement that I could use? Here I provide you some of my own micrographs 2D templates, my Ab-initio vs what it should look like.
Any advice and comment would be much appreciated. Thank you!
settings: 2D: 50 classes, Initial classification uncertainty factor 6,separation distance 10, final full iterations 5, number of final full iterations 50, batch-size per class 1000
ab initio: max resolution 4, initial resolution 15, initial iterations 200, final iterations 300, initial minibatch size 300, final minibatch size 1000, C1 symmetry.
Hetero refinement: batch size per class 1000, learning rate during randomization 0.2, learning rate 0.1, initial resolution 7, max alignment 3, number of initial random assignment iterations 10, number of final full iterations 90