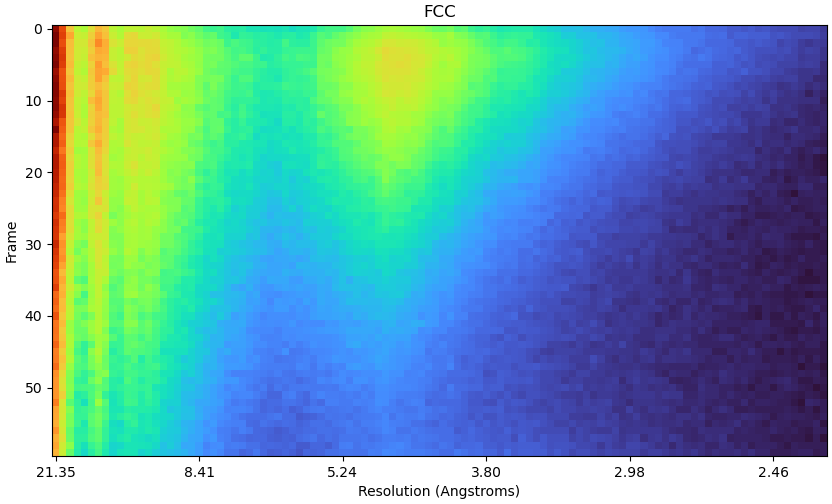
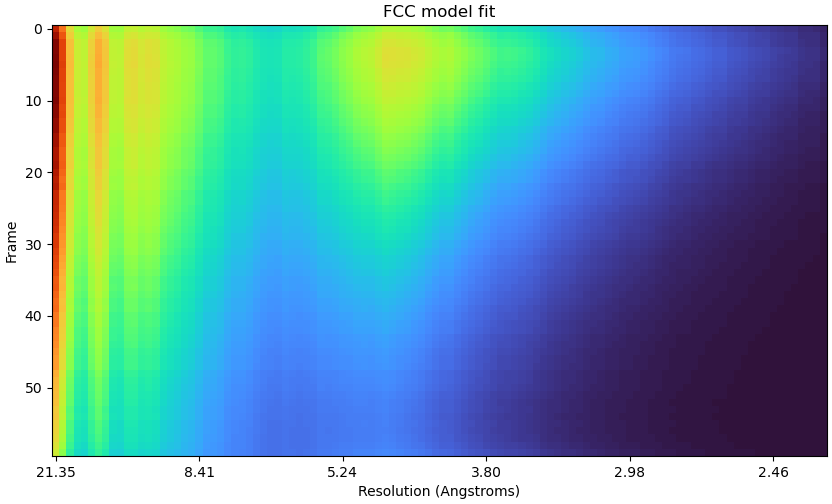
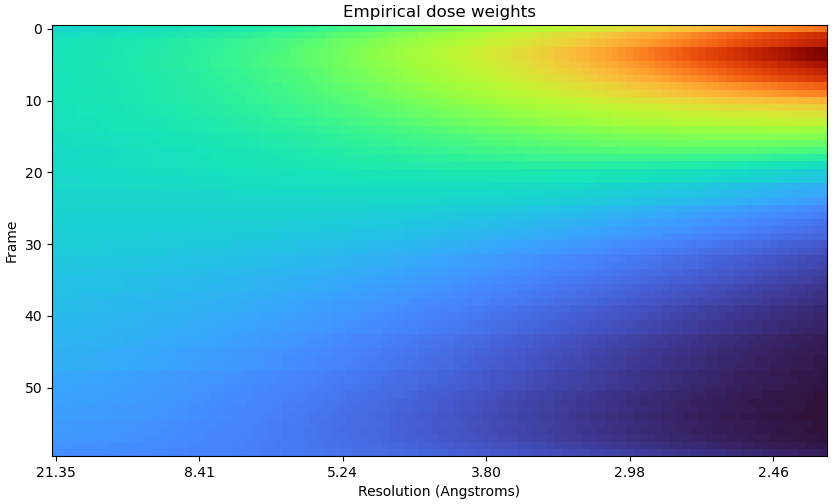
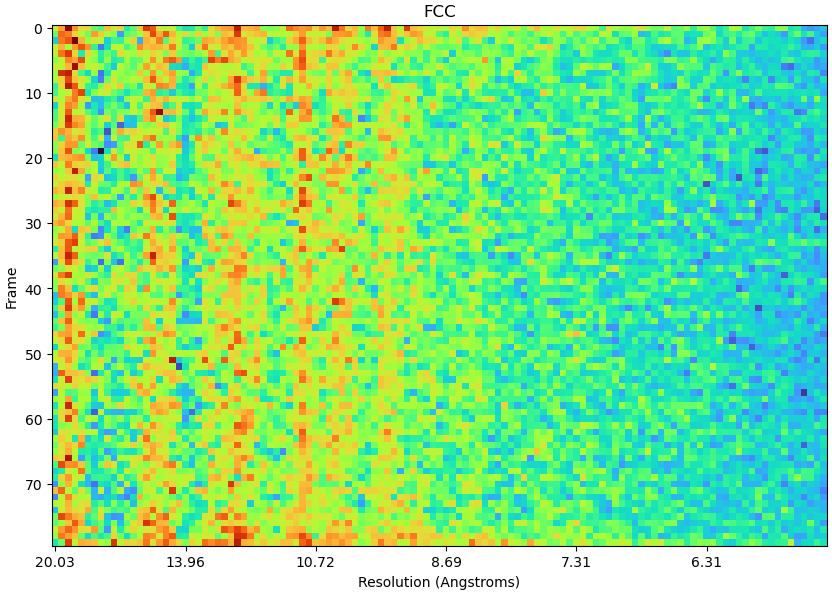
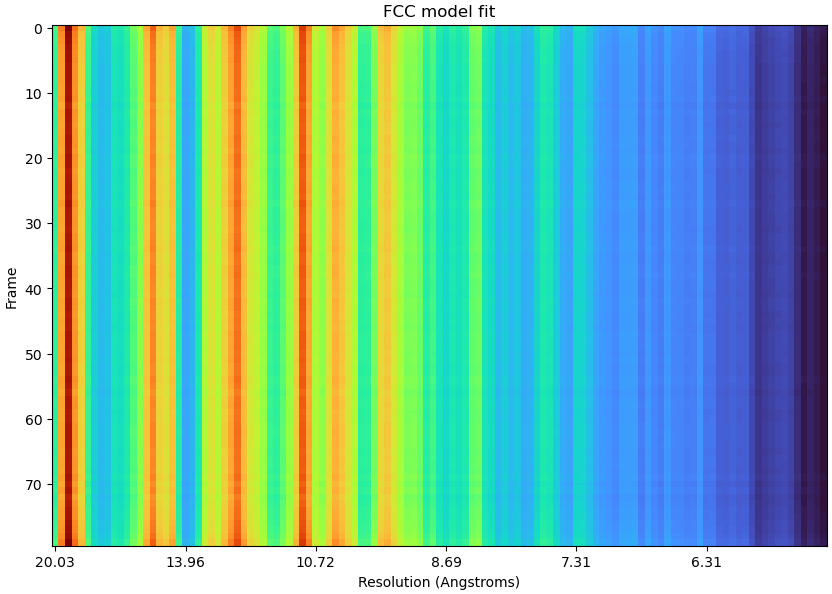
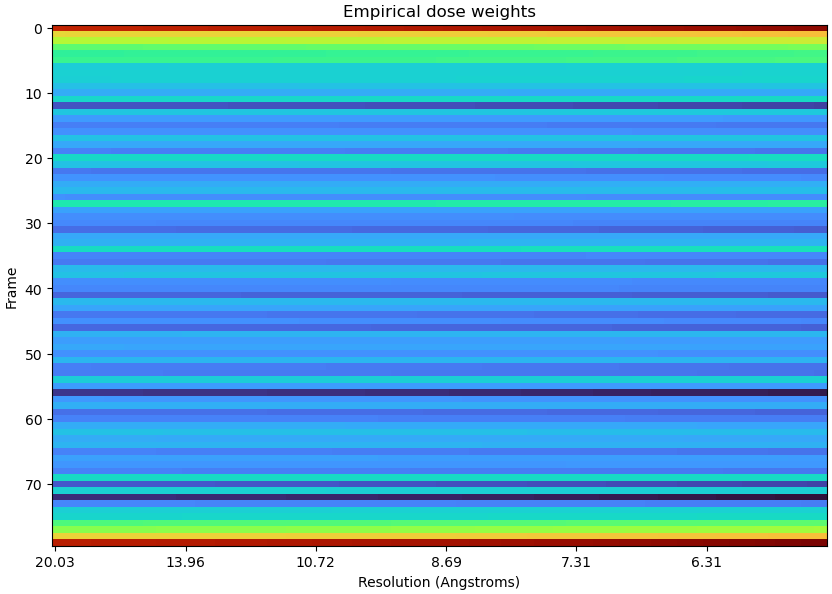
Hi CryoSPARC team,
Had RBMC crash in a new way:
Resolution cutoffs: alignment 3.252 A, cross-validation 2.299 A
Removed 875 movies with fewer than 2 particles.
Recentering particles based on their aligned 3D poses...
Removing 96 particles too close to micrograph edges
--------------------------------------------------------------
STARTING: OPTIMIZE HYPERPARAMETERS
--------------------------------------------------------------
Working with 1137 movies containing 25012 particles
Computing intended data cache configuration
SEARCH RANGES:
zs:
4.6052
6.2146
8.0064
9.7981
thetas:
-1.9373
-2.0420
-2.1468
-2.2515
-2.3562
-2.4609
-2.5656
-2.6704
-2.7751
r start:
0.1000
r end:
10.0000
r step:
0.4950
==================== BEGINNING ITERATION 1 ====================
Iteration overview (parameters to be tried):
---r--- -theta- ---z--- | -spatial- -dist.- --accel--
0.100 -1.937 4.605 | 9.65e-01 100 9.11e-01
0.100 -2.042 4.605 | 9.56e-01 100 9.15e-01
0.100 -2.147 4.605 | 9.47e-01 100 9.20e-01
0.100 -2.251 4.605 | 9.39e-01 100 9.25e-01
0.100 -2.356 4.605 | 9.32e-01 100 9.32e-01
0.100 -2.461 4.605 | 9.25e-01 100 9.39e-01
0.100 -2.566 4.605 | 9.20e-01 100 9.47e-01
0.100 -2.670 4.605 | 9.15e-01 100 9.56e-01
0.100 -2.775 4.605 | 9.11e-01 100 9.65e-01
0.100 -1.937 6.215 | 9.65e-01 500 9.11e-01
0.100 -2.042 6.215 | 9.56e-01 500 9.15e-01
0.100 -2.147 6.215 | 9.47e-01 500 9.20e-01
0.100 -2.251 6.215 | 9.39e-01 500 9.25e-01
0.100 -2.356 6.215 | 9.32e-01 500 9.32e-01
0.100 -2.461 6.215 | 9.25e-01 500 9.39e-01
0.100 -2.566 6.215 | 9.20e-01 500 9.47e-01
0.100 -2.670 6.215 | 9.15e-01 500 9.56e-01
0.100 -2.775 6.215 | 9.11e-01 500 9.65e-01
0.100 -1.937 8.006 | 9.65e-01 3000 9.11e-01
0.100 -2.042 8.006 | 9.56e-01 3000 9.15e-01
0.100 -2.147 8.006 | 9.47e-01 3000 9.20e-01
0.100 -2.251 8.006 | 9.39e-01 3000 9.25e-01
0.100 -2.356 8.006 | 9.32e-01 3000 9.32e-01
0.100 -2.461 8.006 | 9.25e-01 3000 9.39e-01
0.100 -2.566 8.006 | 9.20e-01 3000 9.47e-01
0.100 -2.670 8.006 | 9.15e-01 3000 9.56e-01
0.100 -2.775 8.006 | 9.11e-01 3000 9.65e-01
0.100 -1.937 9.798 | 9.65e-01 18000 9.11e-01
0.100 -2.042 9.798 | 9.56e-01 18000 9.15e-01
0.100 -2.147 9.798 | 9.47e-01 18000 9.20e-01
0.100 -2.251 9.798 | 9.39e-01 18000 9.25e-01
0.100 -2.356 9.798 | 9.32e-01 18000 9.32e-01
0.100 -2.461 9.798 | 9.25e-01 18000 9.39e-01
0.100 -2.566 9.798 | 9.20e-01 18000 9.47e-01
0.100 -2.670 9.798 | 9.15e-01 18000 9.56e-01
0.100 -2.775 9.798 | 9.11e-01 18000 9.65e-01
Cross-validation scores computed:
[▇▇▇▇▇---------------------------------------------------------------------------] 2728/40932 (7%)
DIE: [refmotion worker 3 (NVIDIA RTX A4000)] fatal error: Specified micrograph has less than two particles.
movie 1095300623923001321: J12/imported/001095300623923001321_FoilHole_28436071_Data_28423640_1_20240529_003121_EER.eer
====== Job process terminated abnormally.
Rerunning now, it’s picked different movies this time (at least it’s reporting a different movie count and particle count, so I hope it won’t happen again).
Will send more info privately if desired.
Thanks,
R