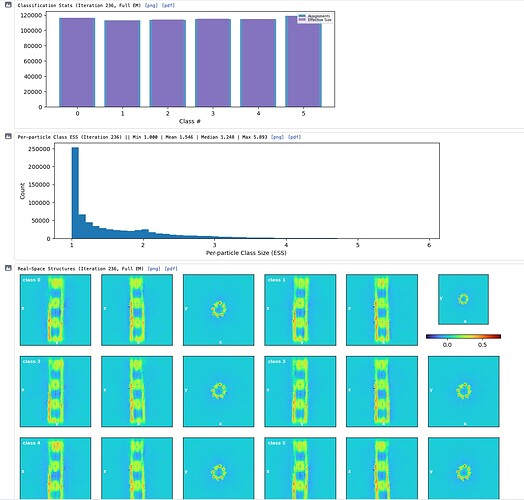
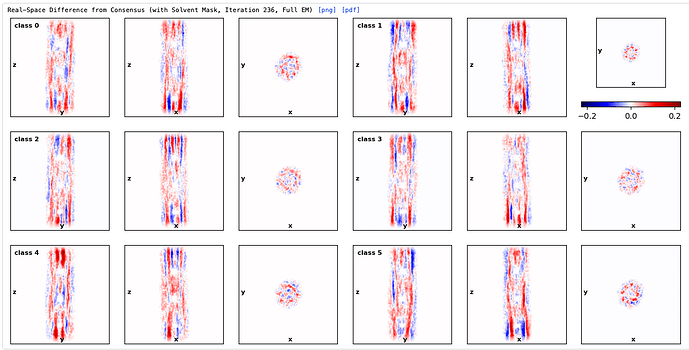
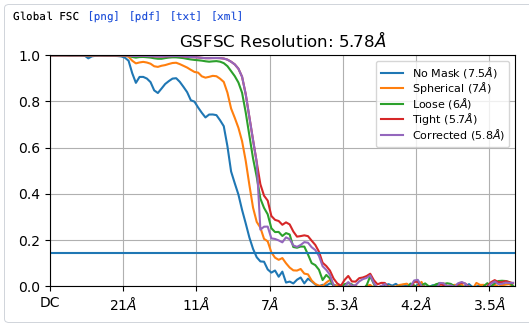
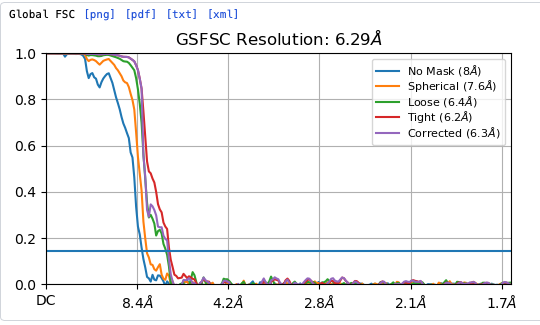
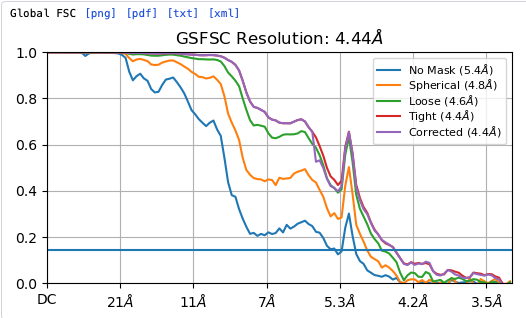
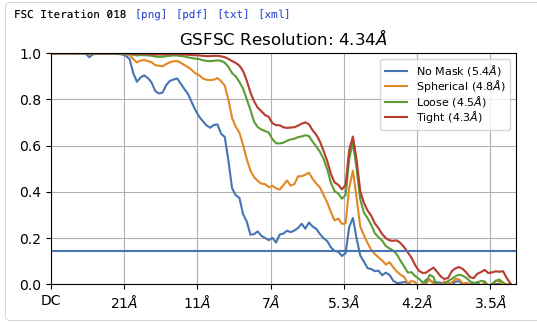
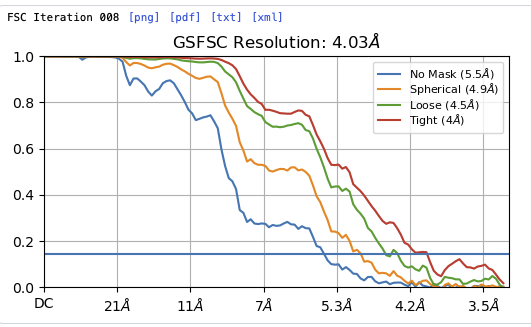
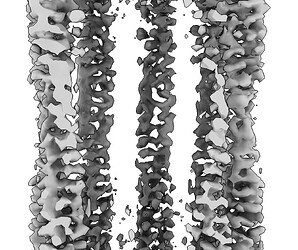
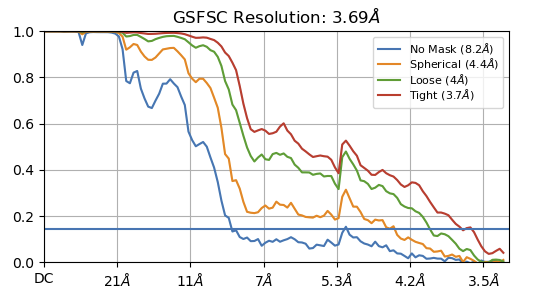
Hey @lorago92 – couldn’t help but jump in here – super fascinating! Based on what you’re describing, perhaps this filament is undergoing a continuous deformation / flexing motion. Depending on how intricate this deformation is, it might make sense that 3D class isn’t able to adequately ‘freeze’ the motion into K consensus volumes.
If you are interested in the relative motion of the three repeated units, I would try:
-
3D Variability Analysis. The motion should be quite clear in one / multiple components. You can try clustering in these components, but I suspect you may run into similar issues as 3D class. The ‘simple’ mode (which decomposes the linear subspace into ‘frames’ along each component) might give you a nice animation of the overall motion. However, in this case, stepping along one of the components may only show part of the whole story. So I would also try:
-
3D Flex – which may help not only visualize this non-trivial continuous heterogeneity, but also ‘undo’ this flexing into a single higher-resolution ‘canonical’ volume (in a similar way to local refinement; which may also be useful here).
P.S., a general note on refinements (quite lengthy, hopefully useful!).
In all three of these jobs we assume that alignments are fixed and proceed to absorb all of the heterogeneity into a more general model of density (i.e., K classes, a linear subspace with K directions, or a non-linear neural-network-based deformation of a canonical volume, respectively). However, in reality, the heterogeneity will be factored into both the map and the alignments – a projection of a continuously deforming structure does not have an ‘alignment’ with respect to a single fixed volume as we usually understand it (i.e., there is no high-res fixed consensus structure and therefore no accurate alignments). In general, the alignments are a ‘nuisance’ parameter – we really care only about the quality of the final map.
As a result, re-refining will not always produce ‘better’ alignments. If one runs classification without alignment, and then re-aligns within a class, the latter step can ‘revert’ the process of the former. This might explain the more ‘diluted’ structures you describe. Since each of the K classes contains multiple states along the continuum, re-refining the alignments assuming a single consensus might reduce the quality of the map.
N.B., iterating classification / alignment can help if there is sufficient signal in each class, and is the basis of classification with alignment (or Heterogeneous Refinement, as we call it). In your case, however, you’d probably need quite a few classes to resolve this motion, which then reduces the signal per class, and probably leads you back to 3DVA / 3DFlex which absorb heterogeneity into a non-static map rather than assume K clusters of consensus structures and associated alignments.