Hi everyone,
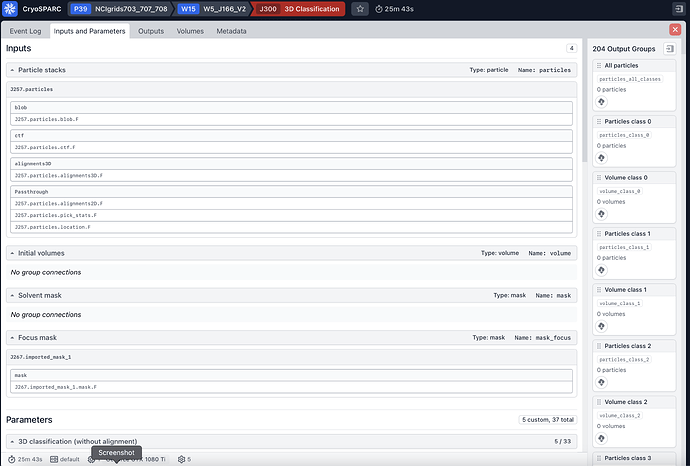
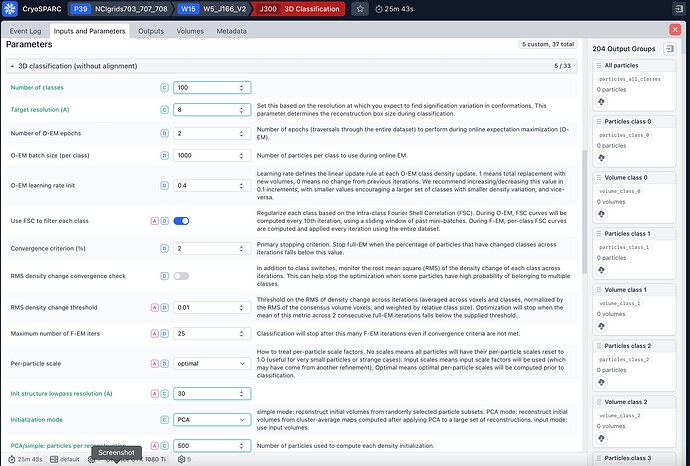
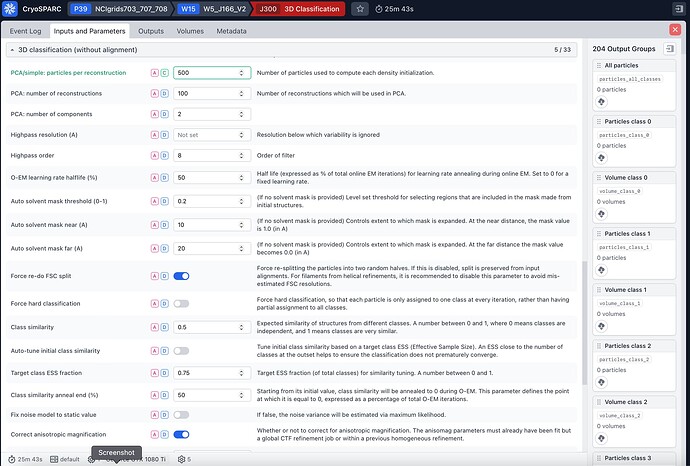
I tried to run a 3D classification job of 740k particles that come from NU-refinement job (input parameters in the screenshots attached), the job gives the following error:
Traceback (most recent call last):
File “cryosparc_master/cryosparc_compute/run.py”, line 96, in cryosparc_compute.run.main
File “cryosparc_master/cryosparc_compute/jobs/class3D/run.py”, line 404, in cryosparc_compute.jobs.class3D.run.run_class_3D
File “/usr/local/cryosparc/2.0/cryosparc2_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.8/site-packages/sklearn/mixture/_base.py”, line 193, in fit
self.fit_predict(X, y)
File “/usr/local/cryosparc/2.0/cryosparc2_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.8/site-packages/sklearn/mixture/_base.py”, line 220, in fit_predict
X = _check_X(X, self.n_components, ensure_min_samples=2)
File “/usr/local/cryosparc/2.0/cryosparc2_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.8/site-packages/sklearn/mixture/_base.py”, line 55, in _check_X
raise ValueError('Expected n_samples >= n_components ’
ValueError: Expected n_samples >= n_components but got n_components = 100, n_samples = 96
My questions are: what does this error mean? Is PCA mode not able to handle a large number of classes like 100?
Thanks in advance.
This happens occasionally to me as well and is fixed by generating 90 PCA classes instead of 100. But 100 also works often, making this frustrating
1 Like
Hi @Proteino (and @CryoEM2),
This error is happening because we do some outlier filtering on the reconstructed volumes prior to clustering. Thus, the 100 original reconstructions (set by the parameter PCA: number of reconstructions) are filtered down to 96. This number is smaller than the number of classes and thus our GMM clustering will fail.
In general, we recommend setting the parameter PCA: number of reconstructions to at least 3-5X the number of classes (e.g., 500 in this case). We’ll make this more clear in a future release!
Also of interest: I recently wrote up a quick explanation of how the PCA mode works here: PCA mode initialization in 3D classification - query - #3 by olibclarke. Note that in our experience, this initialization does not typically produce better volumes than just the ‘simple’ mode (which uses random sample backprojection) but we’re actively exploring how to improve this! We’re happy to hear any feedback for how it does in your case.
Hope that helps,
Valentin
3 Likes
@vperetroukhin @CryoEM2 thank you for your help and explanation
good to know, I never put it together that it errors when I have many classes but don’t change PCA reconstructions.
I will also swap to Simple and see if I experience a noticeable difference and report back. For the record, I always (and necessarily so) use 0.1 Class Similarity, or else nothing gets separated. Typically 8-10Å Target resolution, this gives a result pretty similar to 3DVA clustering which is principally what I use the tool for; broad classification of motion to isolate ~50k particle clusters where conformation/composition are the same to fast-track my pursuit of high resolution of a subset.
3 Likes
I saw your other post as well. there’s quite a bit of this content available in searching the discussion board, from valentin, myself, oli both for old 3D class and the new version. Happy digging! Let us know if you don’t seem to get value from the tool and I’m sure we can tailor some suggestions.
1 Like