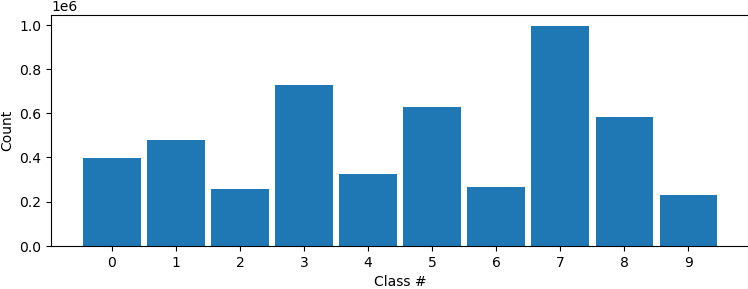
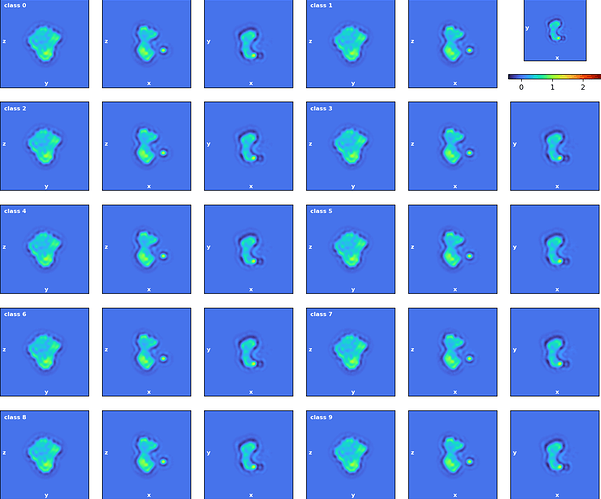
Hi all, I have tried to do a 3Dclass without alignment after running a NU refinement. The histogram showed that some classification may have been made as the particles are not evenly distributed among classes. However, all volumes from intermediate and final classes are the same. Did I do something wrong, or what could be done to show differences after classification? Do I have do heterogenous refinement after 3D classification? Thanks!
Hello @kpsleung,
Are you checking the output volumes directly from the 3D classification job? I would first try Job: Homogeneous Reconstruction Only - CryoSPARC Guide for each class. This generally has helped me better visualizing small differences between classes.
Best,
Kookjoo
I find this job type extremely ineffective, even after homo reconstruction of all volumes which is a huge pain. Port to relion for time being
update: try these settings
O-EM learning rate init - 0.75
O-EM learning rate half-life (iters) - 100
Class similarity - 0.25
let us know if it helps (thanks valentin for suggestion)
although in your case it seems you don’t have strong consensus refinement yet. probably have some work to do upstream first (picking, 2D class selection, etc)
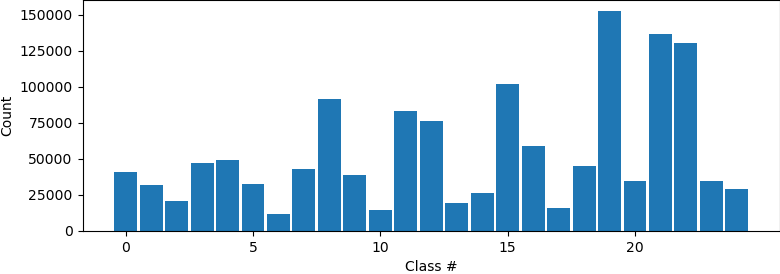
Thank you for your suggestions! I have tried again with the settings, and seems that initial classes can converge into fewer ones:
Default setting:
New setting (intermediate plot):
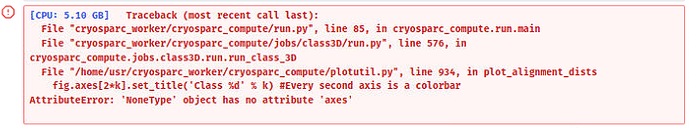
But sometimes the following error hits:
I am now tweaking the parameters to see if classes can be separated with more distinct features / converging into fewer classes, while not hitting any error.
Valentin said this was fixed in a patch. And a new patch came today. Can you update?
Yep! @kpsleung this bug should be fixed in patch 220118 – this will get applied if you update to our newest patch.
@vperetroukhin I can’t wait to try 100 things with the new version. Any magic settings to consider? should defaults work well for small particles or should we try something like this?
I don’t know about magic settings, but “force hard classification” seems to work wonders for cases where ESS was staying very high and all classes were appearing the same.
That, and then tweak the O-EM learning rate if needed (default 0.4; lower gives more diverse classes, so reduce if the defaults cause all particles to collapse into few classes).
The other thing that can make a difference is the target resolution - we usually get the best results from setting it as low as possible while still capturing the desired heterogeneity (unfortunately this can lead to “crunchy” volumes, so I generally resample them in batch using relion_image_handler for inspection).
@CryoEM1 glad you’re excited  ! @olibclarke summarized most of the salient parameters! Check out the new density-based convergence criterion as well – hopefully useful for larger datasets to avoid doing a number of F-EM iterations when the structures themselves have already converged.
! @olibclarke summarized most of the salient parameters! Check out the new density-based convergence criterion as well – hopefully useful for larger datasets to avoid doing a number of F-EM iterations when the structures themselves have already converged.
Another issue I keep running into (processing EMPIAR data) is per-particle scales. Computing these upstream of 3D class can improve the quality of classes quite a bit (see the updated 3D class tutorial for an example).
FYI, we’ve put together a page summarizing some tips on small/membrane proteins in a new tutorial:
Tutorial: Tips for Membrane Protein Structures - CryoSPARC Guide.
Let us know if you have any other useful suggestions!
Hi @vperetroukhin - regarding the density based convergence criteria - how exactly is “RMS Density Change” calculated, and what is a reasonable value to use as a convergence criterion? Also what is “Mean KL Divergence”?
RMS density change: the root-mean-square (mean over voxels) of the real-space density difference across two iterations. We compute this value for each class volume, normalize by the RMS value of the consensus volume density, and then take a weighted average across classes, where the weights are set by the relative class size (this weighted average accounts for classes with tens of particles which can undergo large density changes that are not important). The default value of 0.01 seems to work well with different jobs I’ve tried – in general, setting this might be a bit of a trial and error (e.g., output volumes after every F-EM, track this number in the class flow diagrams and adjust).
Mean KL Divergence: is a ‘soft’ version of class switches. This is computed as (1 / N) \sum_i KLD(curr_i | prev_i), where curr/prev are probability mass functions for each particle i. We currently do not use this for any stopping criterion, but I thought it’d be useful to track when ESS values are higher than 1. For example, this may happen if the class posteriors are multimodal and particles ‘jump’ from one mode to the other without the posterior changing much. If this is the case (i.e., KLD is low, but class switches don’t fall), the RMS criterion may then be useful to turn on.
thank you @vperetroukhin that is a very helpful explanation!
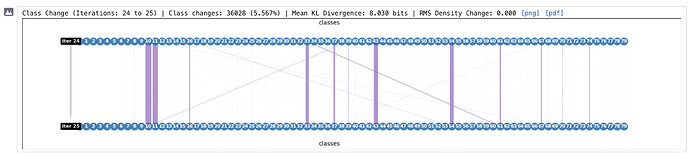
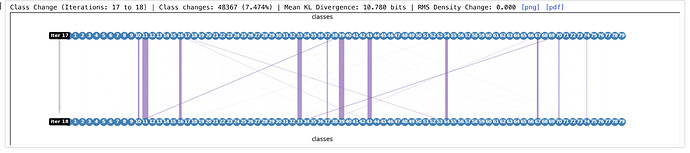
I have a run going at the minute where it has gone through several F-EM cycles and the class switching % is staying stubbornly high (~5%) while RMS density change has gone to zero. If I kill it and mark it as complete, will I be able to use the outputs? Or only if I set it to output results after every F-EM cycle?
Cheers
Oli
No problem! Unfortunately, no – mark as complete won’t let you use those outputs unless you have the F-EM output option set to true.
Gotcha - that’s a shame - it’s a lot of computation and the “good” classes look good! Maybe it might be worth adding a default “floor” of RMS density change of 0.000 for cases like this? Or allow job completion to trigger generation of outputs somehow - or maybe just let users set a maximum number of F-EM iterations, so that it is ensured to terminate at some point?
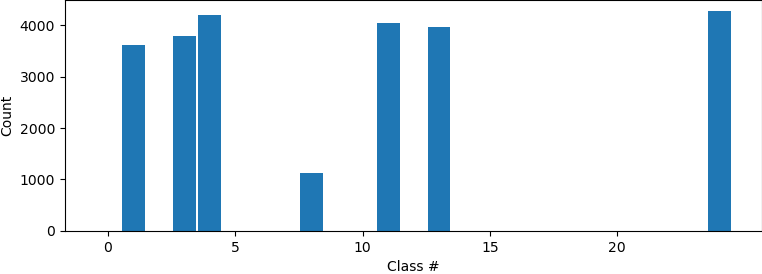
Looking at it further, I wonder if the RMS density change value printed on the graph is correct - it is listed as 0.000 even in the first F-EM iteration where it is printed, while class switching % is still dropping (and I have seen similar for other samples):
This is an interesting case… I haven’t looked at too many cases where I turned on the RMS criterion and set the number of classes to be greater than ~25. I suspect what’s happening is that average across classes is reducing that number below the usual range. The number should be correct but perhaps we can adjust the format to scientific notation.
One other thing I would try in this case is running a homogeneous refinement with per-particle scales turned on, and then re-doing 3D class. I’ve seen this kind of switching behaviour go away after accounting for the scales.
Thanks but scales are already accounted for (or should be - particles originated from NU-refine w/ per particle scale refinement, then were downsampled, then hetero, then 3DClass)
Maybe it would be worth including a histogram of per particle scales somewhere in the log, so that users can make sure whether scales have been refined or not?
Ah ok, got it. Yep, I think we’ll add that (and perhaps a warning if all scales are set to 1) in a future release.