Hello everyone,
I’m working on a protein-RNA complex where a protein dimer binds two RNA molecules (total molecular weight ~ 200kDa). The sample was prepared on C-flat Cu 2/1 holey carbon grids and collected at a pixel size is 0.832 Å, with defocus values from -0.8 um to -2 um. Particles were extracted with a box size of 352 px and rescaled to 128 px for 2D classification.
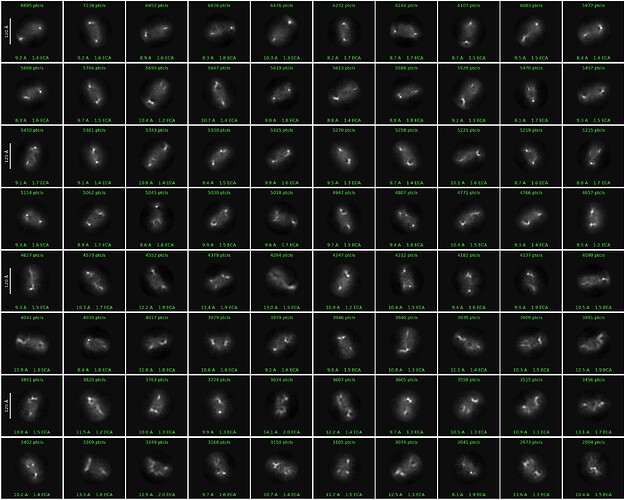
When running 2D classification in Cryopsarc, most 2D class averages consistently show two bright dots at opposite ends of the particles, which likely correspond to the RNA molecules. However, the central protein region appears weak and poorly aligned, resulting in unclear 2D features.
Do you have any suggestion on how to improve the 2D classification in this case?
A representative low pass filtered micrograph and a 2D classification result have been included here.
Thank you for your time and insights!