Hi,
I’m having a weird Corrected FSC curve when processing a particular dataset. Any suggestions of what might be causing this behavior ? This happens for NU and standard protocols.
Regards,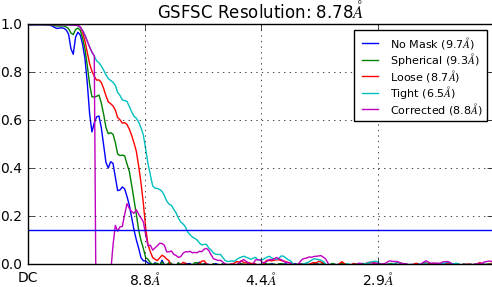
Hi,
I’m having a weird Corrected FSC curve when processing a particular dataset. Any suggestions of what might be causing this behavior ? This happens for NU and standard protocols.
Regards,
Hi @asgodoy,
Thanks for posting.
In this case, did you use a custom mask that was imported in cryoSPARC, or the default “dynamic masking”? how large is your protein, and does the refinement result that you get look reasonable for a 9-10A structure?
Hi @apunjani
We used the default dynamic mask. This is a very elongated protein, almost looking like a filament (size is 250 A x 30 A x 40 A). We dont expect any sort of bending.
TIn the map we are starting to see some 2rd structural elements where they should be.
cheers
Hi @andresgodoy,
Can you try repeating the refinement with the dynamic masking parameters changed:
“Dynamic mask near (A)” : 10A
“Dynamic mask far (A)” : 20A
Please let us know what happens!
I am having a similar issue with this, as my corrected drops fairly quickly and then starts increasing again. See picture below.
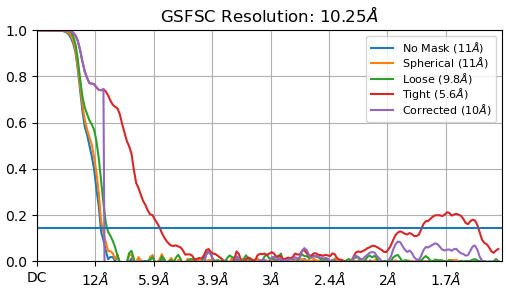
As @andresgodoy said I am starting to see hints of RNA structure, however, I would say that the resolution is slightly overestimated.
Any advice would be much appreciated.
cheers