Hi all,
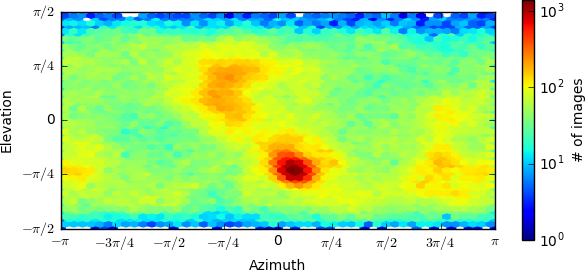
Could someone please help me to figure out whether this map has an
orientation problem? I attached the direction distribution diagram here.
Hi all,
Could someone please help me to figure out whether this map has an
Assuming your map looks like a protein, I would say it has a bias, but not a preference. For some samples this results in distortions in a certain direction in a map, and for others has little effect. It depends on the shape of the protein and the orientation of the secondary structures. For example, in a transmembrane protein composed of parallel alpha helices, side views and top views intrinsically carry different amounts of information.
If the map doesn’t look like a protein, then the particles are probably misaligned and you can’t make any conclusion from the distribution plot, because the orientations are themselves inaccurate. If there is truly a preferred orientation, with no/almost no sampling of other directions, then the map will be so streaky as to be uninterpretable.
This one looks bad, but is actually a pretty decent 3.2 Å structure with no evident distortion.
Yes,this is a prorein particle map.The map is of low quality, and some regions outside have only little density. Do you have any suggestions to improve this map using cryoSPARC or Relion.Thank you very much.
By “looks like a protein” I mean if it’s believable that this map is a real (if low resolution) structure. If there’s a lot of over fitting, then the alignments may be mostly wrong and the map has no interpretation.