hmmmm good question - I have a high dose, small particle dataset somewhere that shows this effect, could try with an unusually large box & see if issue persists
Thought I’d push this discussion back up the list a bit in the hope that more data may lead to resolution with a brief update:
~220 kDa protein dose weighting:
Calculated parameters were:
Using hyperparameters:
Spatial prior strength: 7.3932e-03
Spatial correlation distance: 3000
Acceleration prior strength: 5.2274e-04
…
~300 kDa protein dose weighting:
Calculated parameters were:
Using hyperparameters:
Spatial prior strength: 2.6226e-02
Spatial correlation distance: 18000
Acceleration prior strength: 7.8811e-04
No idea why the plots are different dimensions, both were acquired on the same microscope with the same conditions (EER, 4K rendering, 40 e- total dose, 40 frame split, same magnification). One gets to slightly higher resolution in NU refinement.
One shows excessive weighting of late frames and one doesn’t. Both were estimated with “Extensive” parameter search, same settings (defaults) otherwise on the same system running (I thought?) the same version of CryoSPARC (v4.5.3).
Other than the target sample, the ice conditions in the second dataset (which has good dose weighting) were less than ideal and sample concentration was rather lower than I was happy with, but wasn’t a sample I had any input in at grid prep stage.
Thanks for the additional data point, @rbs_sci. We haven’t forgotten about this. I still don’t have any news to share, but we’re aware that this issue is a barrier for users.
–Harris
Hi @hsnyder - reading the new case study that Hannah just posted it looks like there is an update:
Can you elaborate a little? It seems like this definitely affects the weights in the later frames, but also somewhat in the earlier frames? Have you tested this systematically on other datasets?
Spatial correlation distance seems to be calculated as 500 a lot… I’ve seen it on three datasets very recently, and it jumps out as fairly common (from memory of forum posts).
Given how long it can take on larger boxes, I’m going to sort back through some runs, see if there could be a “good enough” prior set for general use (similar to the defaults RELION provides). I’ll post here (or in another thread?), if others are interested - I’d also be interested in if other users repeatedly see similar numbers.
Perhaps useful info would be:
- prior parameters
- calculation type (fast/balanced/extensive)
- detector
- super res (yes/no)
- file type
- total frames
- total dose
Follow up question to this - if increasing the stringency of the spatial prior improves estimates of the dose weights (or reduces artifacts) - is there any way to estimate the dose weights just using the particle trajectories inferred from patch motion? Or what values for the hyperparameters would correspond to just using these trajectories?
Long post with some numbers in the summary tags. Final resolutions from these datasets range from ~3.5 Ang to ~1.4 Ang. Some datasets are from EMPIAR (e.g. dataset 10) but most are just what have been processed recently and are still attached to CryoSPARC instances.
The only broad pattern I can see is that super res acquisition (even if Fourier cropping during motion correction) usually means higher spatial correlation distance.
Of the below, 8 of them have a spatial prior of 500. One gives two different sets of parameters between two runs.
Summary
Dataset 1
Priors:
- Spatial prior strength: 3.6279e-03
- Spatial correlation distance: 500
- Acceleration prior strength: 2.6015e-02
Calc type: balanced
Detector: Falcon 4i
Super res: no
File type: EER
Total frames: 50
Total dose: 50
Dataset 2
Priors:
- Spatial prior strength: 1.4404e-02
- Spatial correlation distance: 3000
- Acceleration prior strength: 2.4311e-04
Calc type: balanced
Detector: Falcon 4i
Super res: no
File type: EER
Total frames: 50
Total dose: 50
Dataset 3
Priors:
- Spatial prior strength: 4.3118e-03
- Spatial correlation distance: 500
- Acceleration prior strength: 2.2788e-04
Calc type: balanced
Detector: Falcon 4i
Super res: no
File type: EER
Total frames: 50
Total dose: 50
Dataset 4
Priors:
- Spatial prior strength: 9.6809e-03
- Spatial correlation distance: 500
- Acceleration prior strength: 7.9174e-04
Calc type: balanced
Detector: K2
Super res: no
File type: TIF
Total frames: 50
Total dose: 80
Dataset 5a (processed on one system)
Priors:
- Spatial prior strength: 7.3932e-03
- Spatial correlation distance: 500
- Acceleration prior strength: 5.2274e-04
Calc type: extensive
Detector: K3
Super res: no
File type: TIF
Total frames: 40
Total dose: 40
Dataset 5b (reprocessed on a different system of same spec - don’t ask)
Priors:
- Spatial prior strength: 1.2677e-02
- Spatial correlation distance: 3000
- Acceleration prior strength: 1.1992e-03
Calc type: extensive
Detector: K3
Super res: no
File type: TIF
Total frames: 40
Total dose: 40
Dataset 6
Priors:
- Spatial prior strength: 4.8880e-03
- Spatial correlation distance: 3000
- Acceleration prior strength: 4.8880e-03
Calc type: extensive
Detector: Falcon 4 (not i)
Super res: yes
File type: EER
Total frames: 60
Total dose: 40
Dataset 7
Priors:
- Spatial prior strength: 1.7274e-02
- Spatial correlation distance: 18000
- Acceleration prior strength: 1.2645e-01
Calc type: extensive
Detector: Falcon 4i
Super res: yes
File type: EER
Total frames: 70
Total dose: 46
Dataset 8
Priors:
- Spatial prior strength: 9.8435e-03
- Spatial correlation distance: 3000
- Acceleration prior strength: 9.8435e-03
Calc type: extensive
Detector: K2
Super res: no
File type: TIF
Total frames: 20
Total dose: 34
Dataset 9
Priors:
- Spatial prior strength: 1.4229e-02
- Spatial correlation distance: 18000
- Acceleration prior strength: 1.9547e-01
Calc type: extensive
Detector: Falcon 4 (not i)
Super res: yes
File type: EER
Total frames: 75
Total dose: 50
Dataset 10
Priors:
- Spatial prior strength: 6.5870e-03
- Spatial correlation distance: 3000
- Acceleration prior strength: 1.4544e-01
Calc type: balanced
Detector: Falcon 3
Super res: no
File type: TIF
Total frames: 200
Total dose: 30
Dataset 11
Priors:
- Spatial prior strength: 2.6226e-02
- Spatial correlation distance: 18000
- Acceleration prior strength: 7.8811e-04
Calc type: extensive
Detector: Falcon 4i
Super res: yes
File type: EER
Total frames: 40
Total dose: 40
Dataset 12
Priors:
- Spatial prior strength: 2.6226e-02
- Spatial correlation distance: 18000
- Acceleration prior strength: 7.8811e-04
Calc type: extensive
Detector: Falcon 4 (not i)
Super res: yes
File type: EER
Total frames: 40
Total dose: 40
Dataset 13
Priors:
- Spatial prior strength: 3.4444e-03
- Spatial correlation distance: 500
- Acceleration prior strength: 3.4444e-03
Calc type: extensive
Detector: Falcon 4i
Super res: no
File type: EER
Total frames: 60
Total dose: 60
Dataset 14
Priors:
- Spatial prior strength: 4.3118e-03
- Spatial correlation distance: 500
- Acceleration prior strength: 2.2788e-04
Calc type: fast
Detector: Falcon 4i
Super res: no
File type: EER
Total frames: 40
Total dose: 40
Dataset 15
Priors:
- Spatial prior strength: 3.6279e-03
- Spatial correlation distance: 500
- Acceleration prior strength: 2.6015e-02
Calc type: fast
Detector: Falcon 4i
Super res: yes
File type: EER
Total frames: 32
Total dose: 32
Dataset 16
Priors:
- Spatial prior strength: 1.0976e-02
- Spatial correlation distance: 500
- Acceleration prior strength: 5.3389e-02
Calc type: balanced
Detector: K3
Super res: yes
File type: TIF
Total frames: 30
Total dose: 24
In case it’s helpful, here are some observations from datasets I worked on since RBMC was added to CryoSPARC.
Same info as given by @rbs_sci for 6 other datasets
Dataset 1
Priors:
- Spatial prior strength: 6.3103e-03
- Spatial correlation distance: 500
- Acceleration prior strength: 3.7268e-02
Calc type: fast
Detector: Falcon3
Super res: no
File type: MRC (but used as TIFF generated by relion_convert_to_tiff by the facility)
Total frames: 40
Total dose: 40
Dataset 2
Priors:
- Spatial prior strength: 4.6001e-03
- Spatial correlation distance: 500
- Acceleration prior strength: 6.4435e-02
Calc type: fast
Detector: K3 with energy filter
Super res: no
File type: TIFF
Total frames: 40
Total dose: 40
Dataset 3
Priors:
- Spatial prior strength: 3.6279e-03
- Spatial correlation distance: 500
- Acceleration prior strength: 2.6015e-02
Calc type: fast
Detector: K3 with energy filter
Super res: no
File type: TIFF
Total frames: 61
Total dose: 43.74
Dataset 4
Priors:
- Spatial prior strength: 3.6279e-03
- Spatial correlation distance: 500
- Acceleration prior strength: 2.6015e-02
Calc type: fast
Detector: K3 with energy filter
Super res: no
File type: TIFF
Total frames: 40
Total dose: 64.76
Dataset 5
Priors:
- Spatial prior strength: 4.2429e-03
- Spatial correlation distance: 500
- Acceleration prior strength: 1.1760e-03
Calc type: fast
Detector: K3 with energy filter
Super res: no
File type: TIFF
Total frames: 40
Total dose: 64.76
Dataset 6
Priors:
- Spatial prior strength: 4.2429e-03
- Spatial correlation distance: 500
- Acceleration prior strength: 1.1760e-03
Calc type: fast
Detector: K3 with energy filter
Super res: no
File type: TIFF
Total frames: 40
Total dose: 64.76
These are really handy! Might be worth adding particle size & grid type? As presumably ice thickness and gold/carbon will affect things? And maybe particles/mic, as presumably (?) this would affect spatial correlation distance?
Pretty much all our samples default to R1.2/1.3Cu, but I’ll check whether/which ones had carbon support film added as that may also make a difference (I expect more than Cu/Au, to be honest). I’ll look up particle size/count as well but need to focus on other things for a bit. Maximum dimension OK?
I’ll update the post with further info as I can.
Same here, I will look this up and edit my previous message to add this info. It was most likely all Cu 1.2/1.3, but I will double-check.
Hi all, thank you very much for compiling your findings and observations!
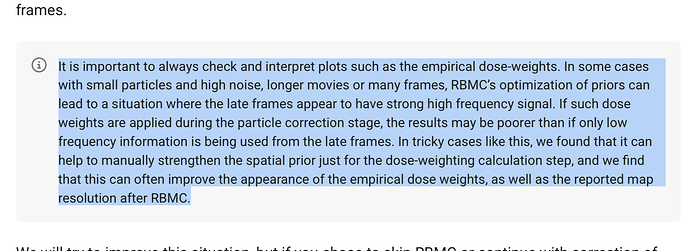
Regarding the effect of strengthening the spatial prior on dose weighting, we have not done a systematic cross-dataset comparison ourselves, so I definitely don’t want to overstate this, but yes, on a few datasets we have seen that strengthening the spatial prior for only the dose weight computation stage can improve final map resolution. We’re still investigating the mechanism behind this behaviour, so I don’t have too much to add at this stage.
Also, note that in RBMC if the search strategy is set to “fast”, 500 is the only value that we actually try for the spatial correlation distance - the search is just 2-dimensional (over the other two params) in “fast” mode.
–Harris
Thanks Harris!!
Re spatial correlation - presumably this will depend mainly on how big and sparsely distributed your particles are. Assuming this is specified in Å, would it be reasonable to do a fast search and set it roughly scaled to the inter particle distance?
I think right now though this is not possible, because you need to specify all three hyperparameters or none at all. But it the spatial correlation is not being searched in fast mode, maybe it would make sense to make it user editable?
@olibclarke the spatial correlation distance is indeed in Angstroms. But, it shouldn’t depend on how sparse the particles are - the whole point of the spatial correlation distance is to set a characteristic scale for the distance over which we expect trajectories to be correlated. It should depend mainly on the physics of the ice deformation (or at least, so we’ve reasoned), which is our justification for leaving it constant in “fast” mode.
That said, you’re right that it might be useful to be able to manually fix just one of the parameters… I’ll make a note of this.
Ah gotcha - yes that makes sense. I guess my thought was that correlation is a matter of degrees, not a binary, and if you have crowded particles vs very sparse ones, the optimal value might be different irrespective of the physics, but maybe that is not the right way to think about it…?
Interesting, thanks Harris. ![]()
Protein won’t move in the same fashion as ice, though, and if severely crowded (something I see more and more in data) then I would suggest that this might change the motion characteristics of the imaged area upon exposure.
This wouldn’t change the broad strokes of the motion (e.g.: doming, vaporisation due to excessive exposure) but may need to be considered if it would change particle motion dramatically in later frames?
Regarding the effect of strengthening the spatial prior on dose weighting, we have not done a systematic cross-dataset comparison ourselves, so I definitely don’t want to overstate this, but yes, on a few datasets we have seen that strengthening the spatial prior for only the dose weight computation stage can improve final map resolution. We’re still investigating the mechanism behind this behaviour, so I don’t have too much to add at this stage.
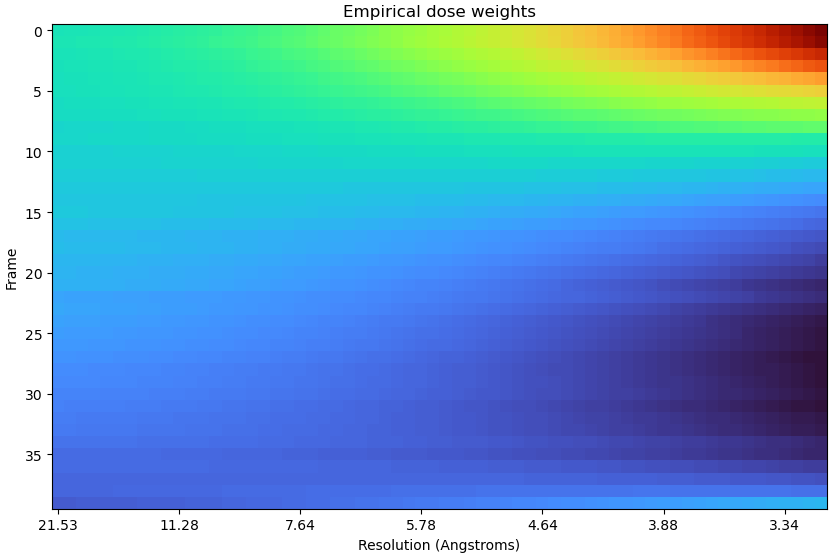
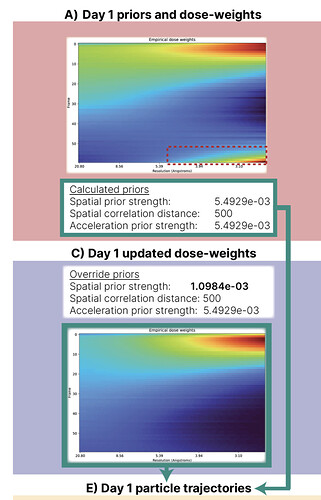
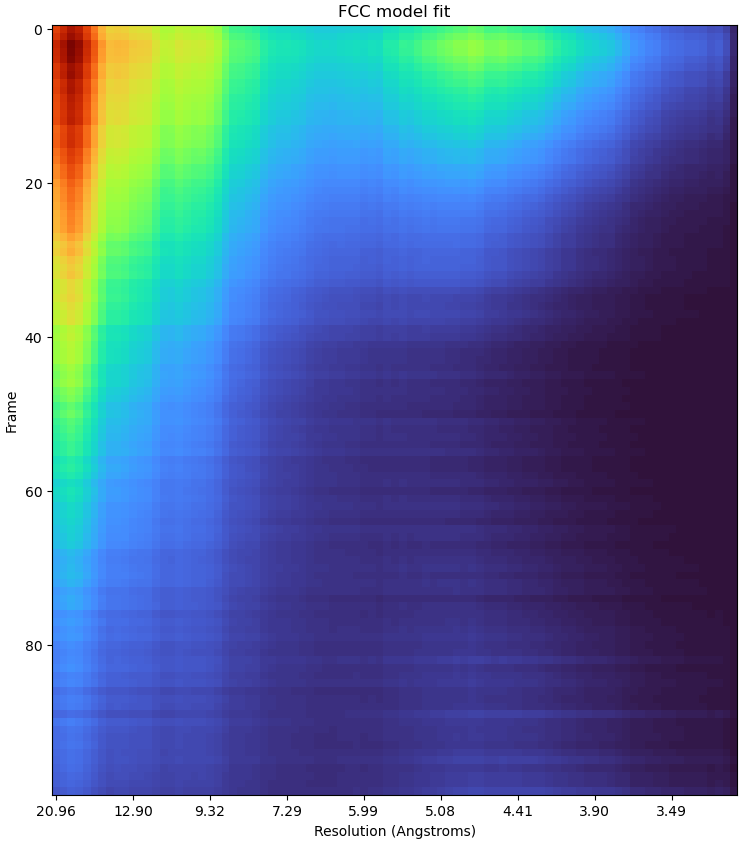
Just to add some more anecdata, I tested this on a 41k subset of the HA trimer dataset and it definitely seemed to help. Dose weights initially looked like this:
Taking this through to motion correction gave a map at 3.2 Å - same as before RBMC.
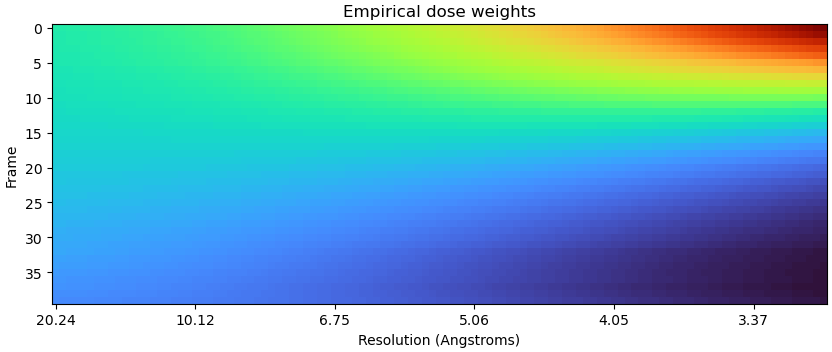
Using a 5x tighter spatial prior for dose weight estimation gave this:
Resulting in a refinement at 3.04 Å.
Using a 10x tighter spatial prior gave this:
Resulting in a 2.99 Å reconstruction.
0.2 Å is not to be sneezed at! If that translates even half as well at higher resolutions (<2 Å) then parameter sensitivity is going to end up needing a much closer look…
Hi Oli
This is really interesting. I was wondering what happens if you just take the per particle trajectories for this data set and keep the original dose weighting from the initial motion correction job.
Thanks
Indrajit
Hi @olibclarke and others,
thanks for this conversation. We have seen this same phenomenon and are trying to change the priors to mitigate the high-spatial frequency components of the later frames.
I’ve seen discussion on changing the spatial prior. Can someone give more detail on how to do this? I see the prior overrides in the RBMC job. Do I pull in the hyper-parameters from the previous job and then set overrides? Do I start a new job from the beginning and add in the overrides? I’m a little confused because there is discussion of “strengthening the spatial prior for only the dose weight computation stage”, but in the RBMC job the only setting under the Empirical Dose-Weighting section are “Use all Fourier components” toggle and “Target number of particles”.
Thanks in advance!
-Rick