I just throw away all the late frames now, say from 30 on or 50 for plots from @rbs_sci and @simonbrown respectively. Then it doesn’t matter.
BTW, I’ve solved a 3.0A structure with < 4e-/A2, so I’m pretty confident you won’t need those late frames anyway.
1 Like
out of interest - global or local searches? (for the 4e-/Å2) & what size particle?
Global searches - actually full “de novo” basic SPA pipeline in cryoSPARC for a ~450 kDa coronavirus spike trimer. (In fact these were the 0˚ images from a cryo-ET test collection).
1 Like
Interesting! In C1? It has to be worse than using a full dose-weighted stack though, no…?
It’s C3. It’s actually not worse than an ~equal number of images/particles. I was surprised!
~1000 micrographs @ 60e-/Å2 → 2.0 Å
~300 of those → 3.0Å
~300 0˚ images from tilt series @ 3.9e-/Å2 → 3.0Å
2 Likes
Huh. Well that is surprising indeed… I guess that probably breaks down for smaller particles, but even so, good to know!
It’s kind of like Wah’s recent preprint where they show worse resolution when you use more tilts (due to radiation damage), except it’s different/stronger because I used all new global searches. Will probably write up a brief preprint at some point.
I also got structures using the whole tilt series - but then finding you could just use the 0˚ ones and actually get a better structure was a little disheartening!
3 Likes
Similar to something I’d been playing with - minimal difference from 60e all the way down to 5e. Below 10e, the challenge becomes seeing particles rather than anything else.
1 Like
I thought it’s still fairly easy with a strong low-pass filter. Definitely less so than high doses. I haven’t tried denoising. (I collected 20 fractions even at @ 3.9 e-/Å2).
At higher defocus also. 5e- was not nightmarish to pick at 2um, but at 0.5um even cryolo was struggling. Still got 2.4A, but 15+ was identifying significantly more particles and consistently hit <2.1A.
These tests at very low dose are super interesting. For sufficiently large particles, one could maybe collect 20 e/A2 instead of the typical 40 (my default), remain safely in the sufficient dose range (given the tests you report with very low dose), and get twice as much data from the same amount of microscope time. Right?
@Guillaume unfortunately not double, because even at high data collection rates > 500/hr only around half the time is spend exposing the camera (very roughly Krios G3 105kx w/ K3 CDS, FPGA binning ~7eps 4s ~40 e-/Å2). Then only 4/3x as much data I think?
@rbs_sci what about estimating CTF in the micrographs? (Using power spectra averages).
1 Like
@Guillaume,
I (fairly) routinely collect lower mag data (250 nm+ viruses) at 20-25 e-/A^2 total dose. Total dose on sensor is usually ~15 e-/A^2 (sometimes lower) due to how thick the ice is and Thon rings are good. Final resolutions are reaching <3 A in optimal conditions, but processing these big boxes is very time consuming (in RELION, CryoSPARC can’t handle 1800+ pixel boxes) and block-based work is also quite slow as I still end up using fairly large boxes and angular sampling converges at very fine sampling.
It’s also interesting how much optimisation can be done for speed. At higher mags, I can break 1,300 micrographs/hour with 1.2/1.3 grids and based on some tinkering earlier this week I’m pretty sure I can break 1,600 with R1/1 and some condition tweaks. Good for smaller things where the ice is thin and very uniform. Not my usual playground…  …so it’s fun.
…so it’s fun.  HexAuFoil is actually a lot slower as the holes are quite far apart.
HexAuFoil is actually a lot slower as the holes are quite far apart.
Data scaling doesn’t work quite that well; the limiting factor is absolutely stage movement, hole alignment and focussing. Breaking 1,000 mics/hr is easy (at least on our Titan w/F4i) but scaling falls off rapidly after that and there is little appreciable speedup in acquisition rate between 40 e- per exposure and 20 e- per exposure (from 1200 to ~1300 on the same grid type and mag). To increase that, decreasing the number of stage movement and focussing steps are the priority - I’m now wondering if 1um holes with 0.5um periodicity would be possible. Also, if the grid was rigid and flat enough, whether you could just focus at the centre of the square and go nuts.
So if entirely in my hands I’d spend a few hours tinkering around with optimal conditions for a given magnification and grid type (I’ve done this for the mags and grids I use the most) but it would do well to make minor adjustments for each sample type.
@DanielAsarnow,
I don’t remember losing an appreciably larger number of micrographs to bad CTF estimation. I normalised the number of micrographs input (500 per dose rate) although that doesn’t account for potential variance in the grid costing me exposures due to less ideal ice conditions.
I do remember significantly fewer picked particles.
I’ll have to check.
1 Like
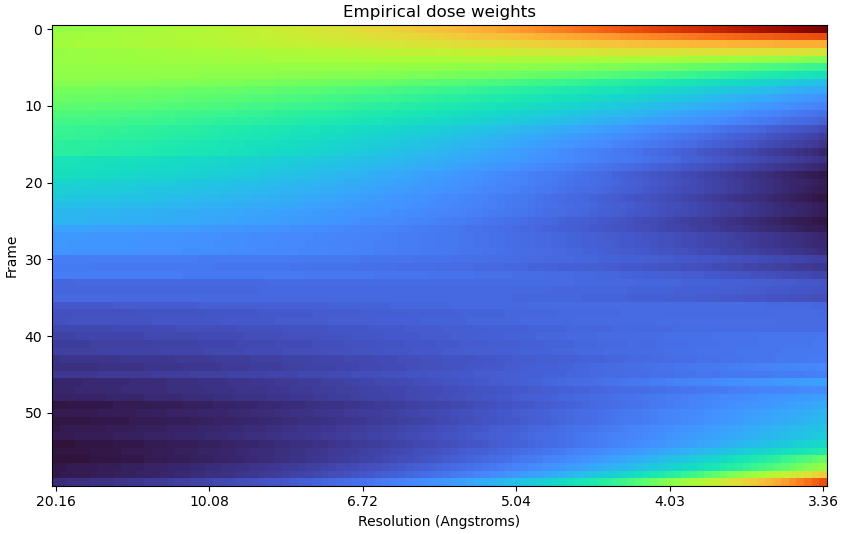
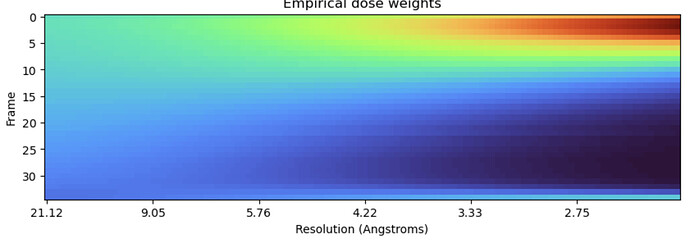
Truncating frames doesn’t always resolve the spurious weightings, at least not completely.
All frames:
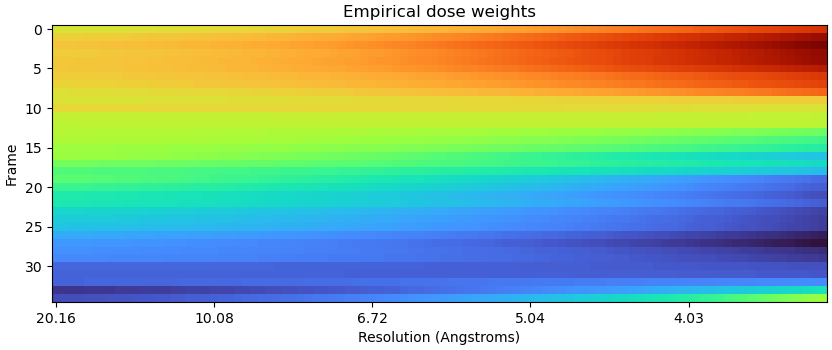
Only first 35 frames:
In this dataset, RBMC failed to improve the resolution until truncating the frames - so I wonder if getting rid of the spurious weights entirely would help even more.
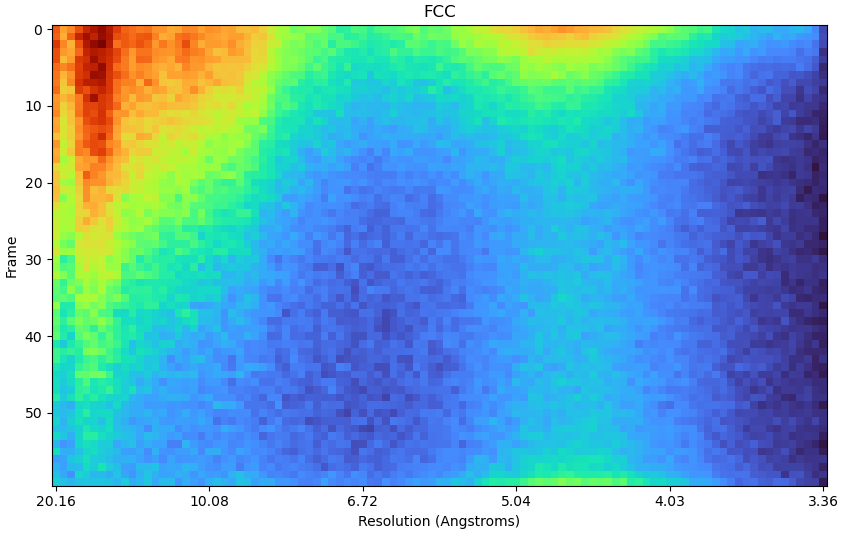
Here is the FCC plot:
1 Like
Thanks @DanielAsarnow for posting these additional results. We’re still tracking this issue but I don’t have any news on it yet.
Hi, @olibclarke ! I’m also dealing with a small protein. I want to improve the total dose from 50 e-/Å^2 to 70 e-/Å^2, how to assess whether the dose is suitable? Can 2D classification on a small subset would be helpful?
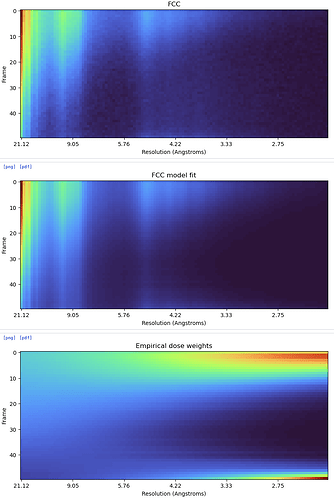
Hi @hsnyder - we still see this even with pretty modest doses (here ~50e-/Å2) - I understand there is no immediate solution to the underlying problem, but it would be useful to have the capacity to choose a frame range for RBMC to avoid having to redo patch motion to mitigate the issue
@olibclarke Truncating the movies doesn’t necessarily fix it as in my last post (though it did allow RBMC to get some resolution improvement). It must be some kind of aliasing.
1 Like
Yes - I’m seeing the same thing… just truncated the previous example to 35 frames, and spurious weightings are still there, if somewhat less pronounced:
That’s why I say it must be some kind of aliasing - given certain combination of factors like spectral resolution, dose rate, box and pixel size (?) there are spurious correlations at a certain resolution range.
These examples with truncation show 1) it appears from nowhere (look at your frame 30 before truncation!) and 2) the total dose doesn’t matter so it’s not a physical effect. So - can we make it go away with parameters we control like the box and pixel size?
And also - since we know per your experiments that it does affect the resolution improvement from RBMC, why not implement a filter that clamps the weights to the generic weighting curve?
1 Like