Hello,
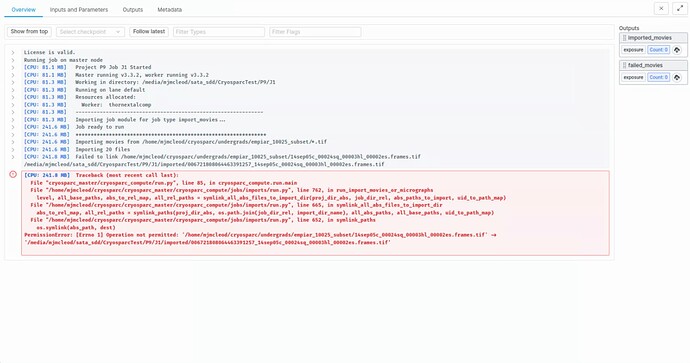
I have been having some trouble with importing micrographs on Cryosparc. Whenever I try to run the import movies job, I get the following error message:
When I navigate to each of the directories shown, cryosparc should have reading and writing permissions, as shown below.
mjmcleod@thornextalcomp:/media/mjmcleod/sata_sdd/CryosparcTest/P9/J1$ ls -l
total 160
-rwxrwxr-x 1 mjmcleod mjmcleod 18 Jun 24 12:57 events.bson
drwxrwxr-x 2 mjmcleod mjmcleod 32768 Jun 24 12:56 gridfs_data
drwxrwxr-x 2 mjmcleod mjmcleod 32768 Jun 24 12:57 imported
-rwxrwxr-x 1 mjmcleod mjmcleod 15088 Jun 24 12:57 job.json
-rwxrwxr-x 1 mjmcleod mjmcleod 583 Jun 24 12:57 job.log
mjmcleod@thornextalcomp:~/cryosparc/undergrads$ ls -l
total 13717728
-rw-rw-r-- 1 mjmcleod mjmcleod 2681112370 Jan 11 01:14 cuda-repo-ubuntu2004-11-6-local_11.6.0-510.39.01-1_amd64.deb
-rw-rw-r-- 1 mjmcleod mjmcleod 2681112370 Jan 11 01:14 cuda-repo-ubuntu2004-11-6-local_11.6.0-510.39.01-1_amd64.deb.1
drwxrwxr-x 2 mjmcleod mjmcleod 4096 Feb 17 2018 empiar_10025_subset
-rw-rw-r-- 1 mjmcleod mjmcleod 8684697600 Jun 22 13:44 empiar_10025_subset.tar
drwxrwxr-x 38 mjmcleod mjmcleod 4096 Jun 21 11:35 P5
I’ve also tried to use the command cryosparcm cli "update_project_directory('P9', '/media/mjmcleod/sata_sdd/CryosparcTest/P9')" , but it gives me the following output:
mjmcleod@thornextalcomp:~$ cryosparcm cli "update_project_directory('P9', '/media/mjmcleod/sata_sdd/CryosparcTest/P9')"
Traceback (most recent call last):
File "/home/mjmcleod/cryosparc/cryosparc_master/deps/anaconda/envs/cryosparc_master_env/lib/python3.7/runpy.py", line 193, in _run_module_as_main
"__main__", mod_spec)
File "/home/mjmcleod/cryosparc/cryosparc_master/deps/anaconda/envs/cryosparc_master_env/lib/python3.7/runpy.py", line 85, in _run_code
exec(code, run_globals)
File "/home/mjmcleod/cryosparc/cryosparc_master/cryosparc_compute/client.py", line 92, in <module>
print(eval("cli."+command))
File "<string>", line 1, in <module>
File "/home/mjmcleod/cryosparc/cryosparc_master/cryosparc_compute/client.py", line 65, in func
assert 'error' not in res, f"Encountered error for method \"{key}\" with params {params}:\n{res['error']['message'] if 'message' in res['error'] else res['error']}"
AssertionError: Encountered error for method "update_project_directory" with params ('P9', '/media/mjmcleod/sata_sdd/CryosparcTest/P9'):
ServerError: Traceback (most recent call last):
File "/home/mjmcleod/cryosparc/cryosparc_master/cryosparc_command/command_core/__init__.py", line 150, in wrapper
res = func(*args, **kwargs)
File "/home/mjmcleod/cryosparc/cryosparc_master/cryosparc_command/command_core/__init__.py", line 3295, in update_project_directory
assert expanded_new_project_dir not in expanded_all_project_dirs, "Project directory %s already exists/used" % expanded_new_project_dir
AssertionError: Project directory /media/mjmcleod/sata_sdd/CryosparcTest/P9 already exists/used
If I make a new folder (in this case titled P9.2) and run the same command, I get the following output:
mjmcleod@thornextalcomp:~$ cryosparcm cli "update_project_directory('P9', '/media/mjmcleod/sata_sdd/CryosparcTest/P9.2')"
None
and the import movies job still returns the same error.
Any help on how to fix this error would be appreciated!