Hello!
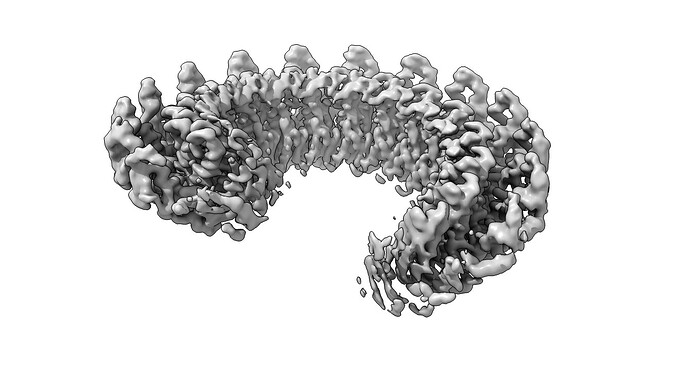
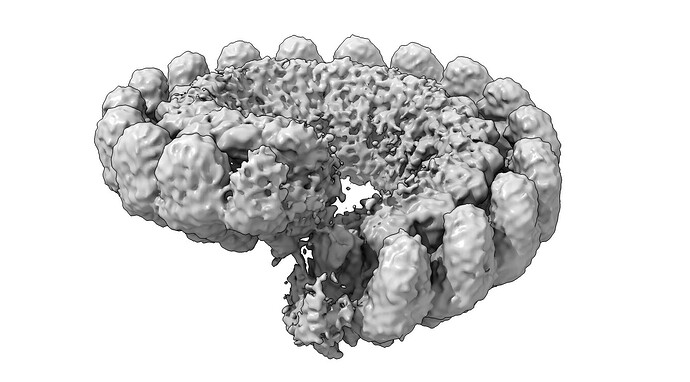
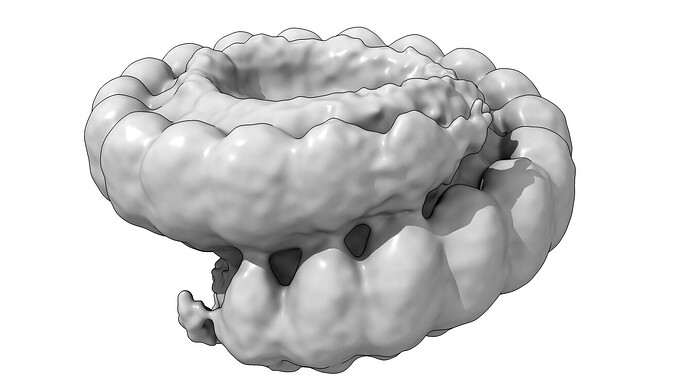
I am puzzled processing one of my states in my dataset. Normally, the protein forms symmetric rings of 19 subunits. However, in the absent of substrate we observe a broken ring structure, that resembles a lock-washer type of assembly.
(another threshold)
However, there is density outside of the normal 19 subunits that we’d expect, which appears to form a continuous helical structure. I am hesitant that the continuous helical state is real, and believe that refinement is misaligning washer projections. The termini of the lock washer seem to be very flexible, causing refinement to focus on more ordered parts.
(gaussian filtered and thresholded to show add’l subunit density)
3DVA display indicated that as density appeared on one termini, the other disappeared, which confirms what i think might be going on in terms of misalignment. I have not been able to separate these particles by 3D classification, either.
My goal is to figure out if my particles are actually forming a continuous filament, or if it is indeed a washer of 19 subunits that cryosparc is shifting during alignment. If it helps, we have MS data that does not indicate any mass species larger than 19-mer.
Thank you,
Carter
1 Like
filaments should be visible in micrographs. with any luck, the 2D would have views that could confirm the subunit count. the 3DVA is a great experiment to see misalignment. Does the problem persist even with perfect references that have no extra density? and providing a mask in NU-refine?
3 Likes
Agree with @CryoEM2; if truly helical, filaments (even if short) should be clearly visible in the micrographs. Think TMV or PyShell, the latter of which occasionally unfolds to a flat sheet.
Could also try mass photometry; been using that to great effect on a couple of controversial (in terms of complex size) samples recently and it’s amazingly sensitive for detecting different multimers!
2 Likes
I would second the suggestions of just inspecting your particles (maybe in the context of denoised mics where contrast is higher) and using mass photometry to compare the mol weight in the presence and absence of substrate.
Also - this may be a case where non-point group symmetry expansion and local refinement, as described here and here, may help to significantly improve resolution and help resolve the intersubunit interfaces.
Separately, you might want to try local refinement around the two ends of the whirl - say the last three or four subunits masked together - and see if that helps clear up what is going on at the broken interface.
2 Likes
Thank you all for the helpful suggestions!
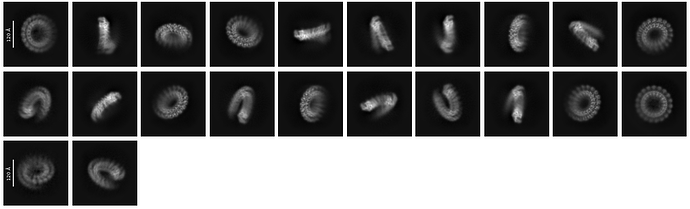
@CryoEM2 @rbs_sci @olibclarke I went ahead and ran a 2D classification of the stack I was working with:
It looks to me like I might have some contamination from ring-shaped particles, as I am not seeing obvious helical assemblies. This could be causing the issue of continuous density (?). This is a little curious to me, since I used 3D classification initially to identify this stack, the same job of which already gave me a class for the 19-mer ring. Does that mean I need to tune 3D class parameters?
We have done a fair bit of mass photometry with this complex, and it seems to discretely form 19-mers. We typically see an equilibrium between monomer and 19-mer, even with substrate. mass spec tells us we have some 18-mers, but we don’t see any with 17,16,15 or so on.
My thoughts are to use heterogenous refinement with reference volumes for washer and ring to separate and then try local refinement with masking of the terminal subunits, but how would you recommend proceeding based on this?
Much appreciated!
Carter
agreed. separate in 2D where obvious beforehand, then het refine with references, or 3D class with references, or 3D class without (0 class similarity) or 3DVA cluster mode. It’s important to remember that ALL data processing jobs in all packages are imperfect, and to varying degrees based on the data. even the 2D above has particles of the wrong identity in each class. So keep classifying iteratively until resolution and homogeneity improve.
2 Likes
I’d also suggest increasing box size about 50%, at least initially. If well centred, the 3D you initially showed can be used as a reference for heterogeneous refinement. I’ve not had a lot of success with using 3DVA to separate discrete states, but that might be a quirk of the samples I’ve tested it on.
1 Like