Hi
We are working with a ~120 kDa elongated complex on graphene oxide grids. We have obtained reasonably good micrographs and can get decent class averages with the gaussian picker or template picker. However, we feel that the background from the GO film and the folds/creases in GO are leading to many false picks.We can of course do multiple rounds of 2D classification to get rid of the false positives but are trying to see if we can use the CryoSPARC Topaz wrapper to do a better job with picking. Unfortunately, we are struggling to generate a good Topaz model and I was wondering if anyone has experience of using Topaz on GO grids. Any suggestions on how we might optimise the model will be really very helpful. I am trying to provide as much details as I can think of below.
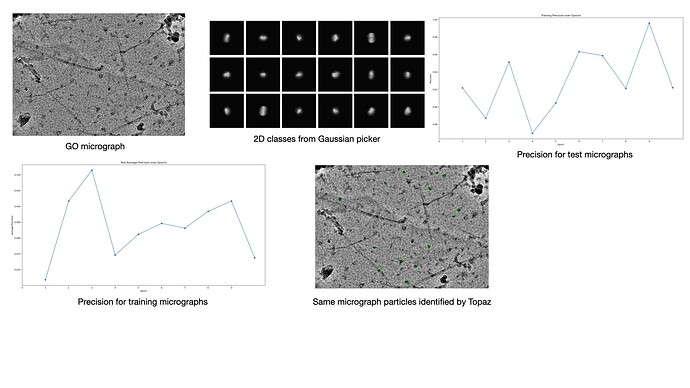
I am attaching a representative micrograph and some 2D class averages obtained with template picking (the particles were heavily binned to a pixel size of 4.2Å/pixel).
I am pasting the Topaz commands from the output log of Topaz train below.
Starting particle pick preprocessing by running command /opt/miniconda/envs/topaz/bin/topaz convert --down-scale 3 --threshold 0 -o /data/Users/Anjali/data_processing/CS-pfaphu/J105/topaz_particles_processed.txt /data/Users/Anjali/data_processing/CS-pfaphu/J105/topaz_particles_raw.txt
Starting dataset splitting by running command /opt/miniconda/envs/topaz/bin/topaz train_test_split --number 11 --seed 1608771774 --image-dir /data/Users/Anjali/data_processing/CS-pfaphu/J105/preprocessed /data/Users/Anjali/data_processing/CS-pfaphu/J105/topaz_particles_processed.txt
Starting training by running command /opt/miniconda/envs/topaz/bin/topaz train --train-images /data/Users/Anjali/data_processing/CS-pfaphu/J105/image_list_train.txt --train-targets /data/Users/Anjali/data_processing/CS-pfaphu/J105/topaz_particles_processed_train.txt --test-images /data/Users/Anjali/data_processing/CS-pfaphu/J105/image_list_test.txt --test-targets /data/Users/Anjali/data_processing/CS-pfaphu/J105/topaz_particles_processed_test.txt --num-particles 100 --learning-rate 0.0002 --minibatch-size 256 --num-epochs 10 --method GE-binomial --slack -1 --autoencoder 0 --l2 0.0 --minibatch-balance 0.0625 --epoch-size 5000 --model resnet8 --units 32 --dropout 0.0 --bn on --unit-scaling 2 --ngf 32 --num-workers 0 --cross-validation-seed 1608771774 --radius 3 --num-particles 100 --device 0 --no-pretrained --save-prefix=/data/Users/Anjali/data_processing/CS-pfaphu/J105/models/model -o /data/Users/Anjali/data_processing/CS-pfaphu/J105/train_test_curve.txt
I am attaching the precision as a fn. of epoch for train and test micrographs
Here is the command from the corresponding Topaz extract job.
Starting extraction by running command /opt/miniconda/envs/topaz/bin/topaz extract --radius 24 --threshold -6 --up-scale 3 --assignment-radius -1 --min-radius 5 --max-radius 100 --step-radius 5 --num-workers 0 --device 0 --model /data/Users/Anjali/data_processing/CS-pfaphu/J105/models/model_epoch03.sav -o /data/Users/Anjali/data_processing/CS-pfaphu/J108/topaz_particles_prediction.txt [58 MICROGRAPH PATHS EXCLUDED FOR LEGIBILITY]
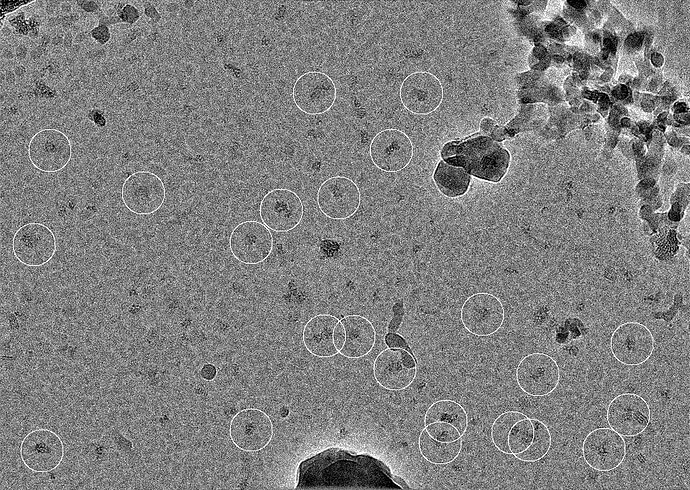
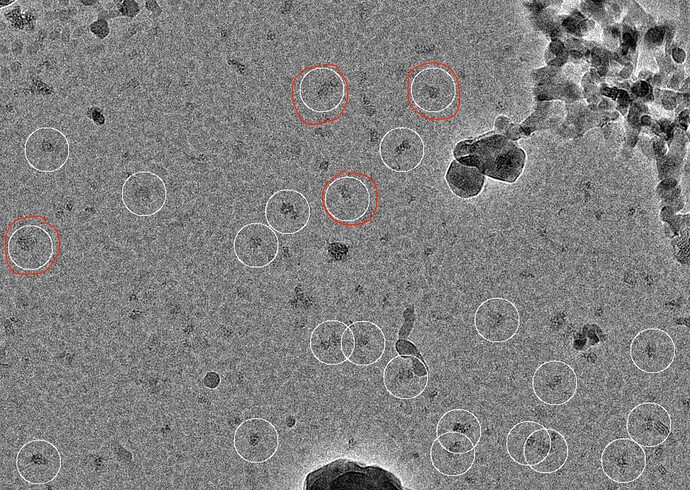
We tested the extraction on a small subset. I am attaching a micrograph with the extracted particles highlighted and as can be seen, while there were very few incorrect picks, many particles were not selected. We also tried using the pre-trained mode of Topaz and with that behaved similar to our template picking job (i.e. we could pick all the particles along with a lot of false positives).