Hello, everyone! I wonder if is is possible for me to extract particles in Cryosparc using those coordinates from Relion.
For example, I extract particles in Relion and get A_particles.star; then I get B_particles.star after multi-rounds 2D classification. After that, I import micrographs into Cryosparc . How to extract particles using B_particles.star in Cryosparc ?
The reason why I try to do that is I want to update binning factor in Cryosparc directly with small number of particles.
Hi @Verdandy,
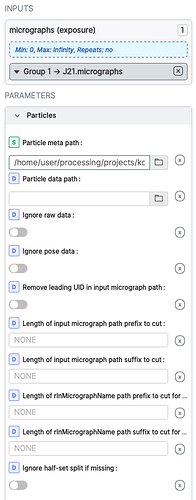
After selecting good classes from Class2D, you can import particles using “Import Particle Stack” job. Assuming you have already imported your movies/aligned micrographs to cryosparc, connect your aligned micrographs as an input group to the newly created “Import Particle Stack” job, and specify the path to B_particles.star file in the particle meta path. This will connect the particle coordinates to your micrographs in the output particle group after this import.
You might have to specify lengths of input or rlnMicrographName path prefix to cut during search, since cryosparc adds some micrograph UIDs in front of movie filenames when the movies are aligned in cryosparc. Normally, I look for the error messages causing the search to fail and fix/edit the parameters accordingly.
After the particle coordinates are imported, I would check how extraction goes with a small number of mics to extract (you can specify this parameter in the export job). You can change binning factors by changing the Fourier crop pixel size in extraction. Hope this helps!
Best,
Kookjoo
Welcome to the forum @Verdandy and thank you for your question.
Thank you @kookjookeem for this response.
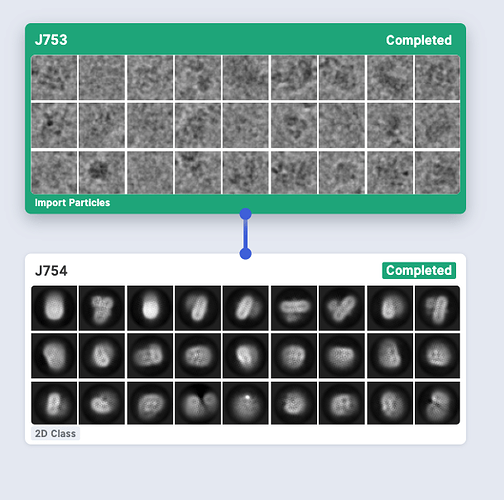
Thanks @kookjookeem and @wtempel , I have connected my import particles and micrographs successfully following your suggestions. But the 2D results seem quite strange without any signals as you can see. I don’t know what is going on.
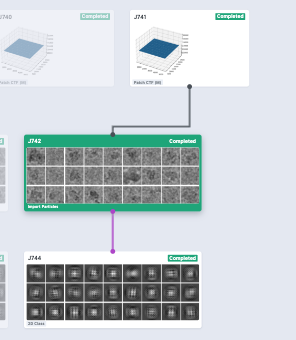
Are the particles extracted with Relion after Class2D? It looks like the “Import Particle” job imported particle coordinates as well as particle images. Could you make sure if the B_particles.star is pointing the right images in _rlnImageName column?
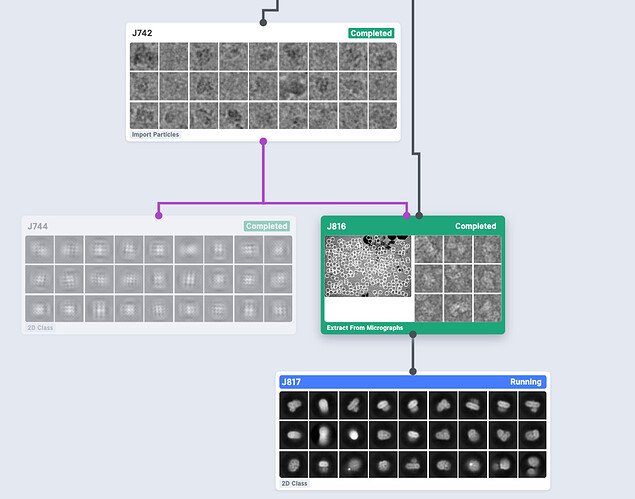
I would also check if the imported coordinates are actually pointing to raw particles on your micrographs (J741) in “Manual Picker”. If the coordinates look fine, I’d run a new extraction using the imported coordinates (all within cryosparc) then run 2D Classification to check if you still have the particle stack with right coordinates.
Best,
Kookjoo
Thanks @kookjookeem, I firstly checked the connection between imported particles and import micrographs and it seemed to be ok from the log.
Then I got normal 2D Classification result using the imported particles which were not connected to micrographs.
Finally, the result of 2D Classification became normal after re-extracting particles. I really don’t know what’s the difference between those particles connected to mics and re-extracted.