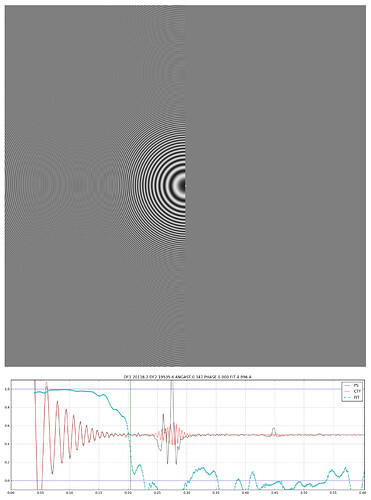
Hi,
In recent versions of cryoSPARC, there seems to be an issue with the display of power spectra in the “individual” view of exposure curation. It seems like they are not correctly normalized or something. You can see an example here - the 1D fit looks okay, but no Thon rings are visible in the raw power spectrum (whereas they are visible with CTFFIND, Gctf etc).
Cheers
Oli
Had the same, and though it was a serious issue with my micrographs. with gctf worked as you said. cheers
Hi @olibclarke,
Do the plots look abnormal in the Patch CTF job itself? Or only in Exposure Curation?
I don’t think these plots are generated during Patch CTF?
I can only see 1D plots (and the “landscape” plot showing local defocus variation) in the log file.
Cheers
Oli
Hi @olibclarke,
When did you notice this happening? Are you using and extra parameters?
Hi @stephan,
Not sure exactly which version this started happening in, but it happens every time, even with default parameters during Patch CTF.
Cheers
Oli
Hi @olibclarke,
Does this happen with all types of datasets?
Ah! Actually now that you mention it, I think it only happens with K3 datasets, K2 datasets look normal…
@marino-j were your misbehaving mics K3 also?
K2 dataset, where defect pixel file is needed…
Also @stephan - the Thon rings in the CTF report for the same dataset in Live look fine, if that’s of any help
Cheers
Oli
1 Like
Hi @olibclarke @marino-j this is really strange - we can’t spot the issue or reproduce yet. As far as we can tell though, the plot being blank does not actually affect the fit CTF parameters or outputs of the patch CTF job - it’s only happening during visualization
OK, thanks a lot for your answer. My way round is to use motioncorr with the defect pixel file, and after that use GCTF. If you need some of these files I can provide them.
Cheers
Jacopo
I have this error as well, I didn’t realize it was a bug until reading this just now.